Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination - PubMed (original) (raw)
Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination
Eric B Dammer et al. PLoS One. 2012.
Abstract
TAR DNA-binding protein 43 (TDP-43) is a major component within ubiquitin-positive inclusions of a number of neurodegenerative diseases that increasingly are considered as TDP-43 proteinopathies. Identities of other inclusion proteins associated with TDP-43 aggregation remain poorly defined. In this study, we identify and quantitate 35 co-aggregating proteins in the detergent-resistant fraction of HEK-293 cells in which TDP-43 or a particularly aggregate prone variant, TDP-S6, were enriched following overexpression, using stable isotope-labeled (SILAC) internal standards and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). We also searched for differential post-translational modification (PTM) sites of ubiquitination. Four sites of ubiquitin conjugation to TDP-43 or TDP-S6 were confirmed by dialkylated GST-TDP-43 external reference peptides, occurring on or near RNA binding motif (RRM) 1. RRM-containing proteins co-enriched in cytoplasmic granular structures in HEK-293 cells and primary motor neurons with insoluble TDP-S6, including cytoplasmic stress granule associated proteins G3BP, PABPC1, and eIF4A1. Proteomic evidence for TDP-43 co-aggregation with paraspeckle markers RBM14, PSF and NonO was also validated by western blot and by immunocytochemistry in HEK-293 cells. An increase in peptides from methylated arginine-glycine-glycine (RGG) RNA-binding motifs of FUS/TLS and hnRNPs was found in the detergent-insoluble fraction of TDP-overexpressing cells. Finally, TDP-43 and TDP-S6 detergent-insoluble species were reduced by mutagenesis of the identified ubiquitination sites, even following oxidative or proteolytic stress. Together, these findings define some of the aggregation partners of TDP-43, and suggest that TDP-43 ubiquitination influences TDP-43 oligomerization.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
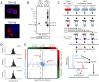
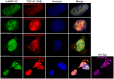
Figure 1. Characterization of the aggregate proteome of HEK-293 cells overexpressing TDP-43 and aggregate prone TDP-S6.
(A) HEK-293 cells were transfected with HA-TDP-43 or HA-TDP-S6 and immunostained with anti-HA antibody for recombinant TDP-43 exclusively (red) and Hoescht stain for the nucleus (blue). Scale bars, 10 µm. (B) Western blotting (WB) of native TDP-43 in the detergent soluble (RIPA) and insoluble (urea) fractions from mock, TDP-43 and TDP-S6 transfected HEK-293 cells. 10 µg and 5 µg of detergent-soluble and insoluble sample were loaded, respectively. A ponceau S nonspecific band was used to show equal loading. (C) Workflow for quantitative proteomics using isotopic labeled internal standards. In this approach, detergent insoluble urea extracts from isotopically-labeled control HEK-293 cells are mixed equally with urea extracts prepared from mock, TDP-43 and TDP-S6 transfected cells. The samples are resolved on an SDS-PAGE gel, excised into gel slices, digested with trypsin, and analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) on a high-resolution Orbitrap mass spectrometer. The isotopically labeled peptides from the control HEK-293 cells are chemically identical to their unlabeled counterparts and serve as internal standards for the measurement of protein abundance across samples. (D) Histograms of the entire population of quantified proteins expressed as log2(light/heavy) vs. frequency for the three mixtures with Mock (null experiment), TDP-43, and TDP-S6. (E) To visualize proteins significantly changing in the TDP-43 and/or TDP-S6 models, we constructed a triple SILAC map of the differences in log2-transformed ratios. Confirming high transfection efficiency, we identified both TDP-43 and TDP-S6 as the most enriched protein component in the cell model (at the top right).
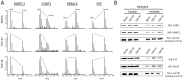
Figure 2. Validation of multiple RNA-binding proteins enriched within the insoluble proteome of TDP-43 and TDP-S6 transfected HEK-293 cells.
(A) Representative quantified peptides from PABPC1, G3BP, RBM14, and PSF in the mock-transfected, TDP-43- and TDP-S6-ovexpressing detergent insoluble HEK-293 proteome with paired internal standard. (B) Western blot (WB) analysis for, PABPC1, G3BP, PSF and NonO in the detergent soluble (10 µg) and detergent insoluble (5 µg) proteome of transfected HEK-293 cells. A non-specific band is shown as a control to confirm equal loading.
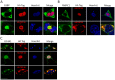
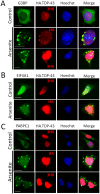
Figure 3. Subcellular localization of stress granule proteins coaggregating with TDP-S6 in HEK-293 cells.
Immunofluorecence confocal microscopy analysis of overexpressed HA.TDP-43 or HA.TDP-S6 (red) with endogenous G3BP (A), PABPC1 (B), or eIF4A1 (C) (green) in HEK-293 cells. Hoechst stain for the nucleus is shown in blue. In panel C, “T” indicates a transfected cell and “U,” untransfected. Scale bars, 10 µm.
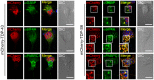
Figure 4. Subcellular localization of stress granule proteins coaggregating with TDP-S6 in primary motor neurons.
Left panel shows localization of G3BP, eIF4A, and PABPC1 (green) in mCherry-TDP-43 overexpressing mouse motor neurons. Right panel, colocalization of the same stress granule markers with mCherry-TDP-S6 in neuronal soma. Nuclei are stained with Hoechst dye (blue). Scale bars, 10 µm.
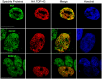
Figure 5. hnRNP A0 colocalization in inclusion bodies positive for overexpressed TDP-43 and TDP-S6 in HEK-293 cells.
Endogenous or overexpressed TDP-43 or TDP-S6 (red) localization relative to endogenous hnRNP A0 (green). In the lower set of images, triple staining was performed to differentiate overexpressed TDP-S6 from endogenous TDP-43 in a neighboring cell; HA staining not included in the merged image is shown in magenta. Nuclei are stained with Hoechst dye (blue). Scale bars, 10 µm.
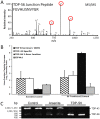
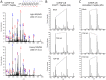
Figure 6. Quantification of detergent insoluble TDP-S6 and TDP-43 following arsenite treatment via LC-MS/MS monitoring of the alternative splicing exon-junction specific TDP-S6 peptide and RT-PCR determination of splicing preference in arsenite-treated HEK-293 cells.
(A) A representative MS/MS fragmentation spectrum of TDP-S6 exon junction peptide F276GVHLISNVYGR, where FG is encoded by the last two codons of a shortened exon 6 in TDP-S6 and the remainder is encoded by an alternative exon composed of bases usually found in the 3′ UTR of TDP-43. The three most intense MS/MS fragment ions on our platform, y6, y7 and y8 are circled in red. (B) The summed intensity of the ions circled in (A) was used to quantify TDP-S6 as described in the methods for Targeted LC-MS/MS. Similarly, a TDP-43 specific peptide (F276GGNPGGFGNQGGFGNSR), and a shared RRM1 peptide F152TEYETQVK were quantified in arsenite-treated HEK-293 detergent insoluble fraction relative to untransfected, untreated (control) cells. Quantification of the same peptides from the detergent insoluble fraction of TDP-S6 transfected cells were measured as a positive control. EIF4A1 was also monitored (via peptide D194QIYDIFQK) to show co-enrichment of a stress granule marker with arsenite treatment or TDP-S6 transfection. Error bars show the 95 percent confidence interval for two replicates. Arsenite treatment (0.5 mM) was for 90 min. (C) Reverse transcriptase (RT)-PCR of TDP-43 and TDP-S6 mRNA species. RT-PCR was performed on total mRNA extracted from biological replicates from control or 0.5 mM arsenite-treated (90 min) HEK-293 cells. Primers selected gave predicted TDP-S6 and TDP-43 specific amplicons of 580 and 1531 bp, respectively. RNA from TDP-S6 transfected cells was used as a positive control.
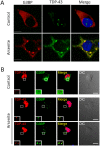
Figure 7. Endogenous TDP-43 participates in arsenite-induced G3BP positive stress granules.
(A) Endogenous TDP-43 (green) stained with an N-terminal monoclonal antibody raised against residues 1–260, and G3BP (red) in control and sodium arsenite (0.5 mM, 45 min)-treated HEK-293 cells. (B) A small fraction of endogenous TDP-43 (red) localizes to G3BP positive (green) cytoplasmic granules induced by sodium arsenite in motor neurons. Nuclei are shown in blue. Scale bars, 10 µm.
Figure 8. Overexpressed TDP-43 participates in arsenite-induced stress granules.
(A–C) Overexpressed TDP-43 (red) localizes to cytoplasmic granules with redistributed G3BP (A), eIF4A1 (B), or PABPC1 (C) (green) induced by arsenite. Nuclei are shown in blue. Scale bars, 10 µm.
Figure 9. Nuclear paraspeckle proteins colocalize with TDP-43.
Immunofluorecence of HA.TDP-43 (red) with endogenous target proteins (RBM14, NonO, or PSF) in green. Hoescht stain for the nucleus is shown in blue in the rightmost panels of each row. Scale bars, 10 µm.
Figure 10. A methylated RGG motif peptide increasing with TDP-43 overexpression identified by unlabeled/SILAC peptide comparison.
(A) MS/MS of an hnRNP A/B dimethylated R332 peptide, both unlabeled (upper panel) and heavy (lower panel). (B) MS spectrum for the peptide in (A) at the LC elution peak from which ion intensities for light and heavy peptides were extracted to calculate Table 4 unnormalized quantified relative levels of the modification in mock, TDP-43, and TDP-S6 paired experiments. Replicate 1 (urea/urea mixture) spectra are shown. (C) A representative unmodified peptide for hnRNP A/B (IFVGGLNPEATEEK), the quantified relative levels of which contributed to the normalization factors in Table 2.
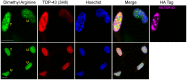
Figure 11. Increased asymmetric dimethylated arginine in nucleus of TDP-43 transfected HEK-293 cells.
Proteins containing asymmetric dimethylarginine (green) were visualized by immunofluorescence in TDP-43 (red)-transfected and -untransfected HEK-293 cells. TDP-43 overexpression in one cell was confirmed by triple labeling also using the antibody recognizing the HA tag (upper row, rightmost panel). Nuclei are shown in blue. “T” indicates a transfected cell and “U,” untransfected. Scale bars, 10 µm.
Figure 12. Identification of ubiquitin sites on TDP-43 by mass spectrometry.
(A) Trypsin digestion of ubiquitin conjugates generates a di-glycine tag (GG), with a monoisotopic mass of 114.0429 Da) on ubiquitin-modified lysine residues, producing unique MS/MS spectra that can be matched by SEQUEST. Searching against a concatenated target-decoy database followed by manual validation, three sites of lysine ubiquitination (K102, K114 and K181) were mapped to TDP-43 in the TDP-S6 insoluble proteome. A fourth site (K145) identified later is also shown. (B) Ubiquitination site context in the linear domain structure of TDP-S6 is shown. (C) Two views rotated about the y-axis of a three-dimensional model of RRM-1 docked to HIV TAR ssDNA (blue) modified from Kuo, et al . K102, K114, K145, and K181 side chains are labeled and explicitly shown (purple) in the context of the RRM-1 domain structure. The closest ε-amino group to the nucleic acid backbone (3.1 Å) was K181, which resides in the loop that follows the RRM1 C-terminus.
Figure 13. Dialkylated peptide standard development for discovery and validation of ubiquitinated lysine peptide MS/MS spectra.
(A) Workflow for generating pseudo-ubiquitinated peptides. Peptides for all spectra generated and detected are listed in Table 5. (B) Comparison of MS/MS fragmentation spectra for the K102 pseudo-GG containing peptide (left panel) and the true K102-GG peptide (right panel). Spectra of all true lysine ubiquitination site peptides, side-by-side with those for pseudo-GG peptides, are given in Table S5.
Figure 14. Biochemical fractionation of TDP-43 and TDP-S6 into detergent soluble and insoluble fractions is altered by the cumulative mutation of the four identified RRM1 lysine ubiquitin attachment sites on TDP-43 and TDP-S6.
(A) Western blot of the N-terminal HA tag (upper panel) on overexpressed TDP-43 or TDP-S6 in HEK-293 extracts indicates a change in high molecular weight species and ubiquitinated species of the overexpressed proteins. Right panels give a lower exposure of the same blot. Expected positions of monoubiquitinated TDP-43 (†), and mono-, di- and tri-ubiquitinated TDP-S6 (*, **, ***) are indicated. 18 µg of RIPA extract and 3 µg of RIPA-insoluble (urea) extracts were loaded. WT, wild type; 4R (K102R/K114R/K145R/K181R); 3R (K102R/K114R/K181R); 2R (K102/K181R for TDP-43 and K114R/K181R for TDP-S6). (B) Detergent-fractionated HEK-293 cell extracts from cells transfected with equal amounts of WT or 4R TDP-43 were immunoblotted for the N-terminal HA tag. Where indicated, cells were treated for 1 h with 0.5 mM sodium arsenite. †, the expected position of monoubiquitinated TDP-43 is indicated. Western blot signal for calnexin (CN, 90 kDa) is provided as a loading control.
Similar articles
- Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains.
Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Seyfried NT, et al. Mol Cell Proteomics. 2010 Apr;9(4):705-18. doi: 10.1074/mcp.M800390-MCP200. Epub 2010 Jan 4. Mol Cell Proteomics. 2010. PMID: 20047951 Free PMC article. - Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards.
Seyfried NT, Gozal YM, Donovan LE, Herskowitz JH, Dammer EB, Xia Q, Ku L, Chang J, Duong DM, Rees HD, Cooper DS, Glass JD, Gearing M, Tansey MG, Lah JJ, Feng Y, Levey AI, Peng J. Seyfried NT, et al. J Proteome Res. 2012 May 4;11(5):2721-38. doi: 10.1021/pr2010814. Epub 2012 Apr 4. J Proteome Res. 2012. PMID: 22416763 Free PMC article. - RNP2 of RNA recognition motif 1 plays a central role in the aberrant modification of TDP-43.
Takagi S, Iguchi Y, Katsuno M, Ishigaki S, Ikenaka K, Fujioka Y, Honda D, Niwa J, Tanaka F, Watanabe H, Adachi H, Sobue G. Takagi S, et al. PLoS One. 2013 Jun 28;8(6):e66966. doi: 10.1371/journal.pone.0066966. Print 2013. PLoS One. 2013. PMID: 23840565 Free PMC article. - TDP-43: a DNA and RNA binding protein with roles in neurodegenerative diseases.
Warraich ST, Yang S, Nicholson GA, Blair IP. Warraich ST, et al. Int J Biochem Cell Biol. 2010 Oct;42(10):1606-9. doi: 10.1016/j.biocel.2010.06.016. Epub 2010 Jun 25. Int J Biochem Cell Biol. 2010. PMID: 20601083 Review. - TDP-43 proteinopathy and mitochondrial abnormalities in neurodegeneration.
Gao J, Wang L, Yan T, Perry G, Wang X. Gao J, et al. Mol Cell Neurosci. 2019 Oct;100:103396. doi: 10.1016/j.mcn.2019.103396. Epub 2019 Aug 21. Mol Cell Neurosci. 2019. PMID: 31445085 Free PMC article. Review.
Cited by
- Quantitative Analysis of the Brain Ubiquitylome in Alzheimer's Disease.
Abreha MH, Dammer EB, Ping L, Zhang T, Duong DM, Gearing M, Lah JJ, Levey AI, Seyfried NT. Abreha MH, et al. Proteomics. 2018 Oct;18(20):e1800108. doi: 10.1002/pmic.201800108. Proteomics. 2018. PMID: 30230243 Free PMC article. - Neuron enriched nuclear proteome isolated from human brain.
Dammer EB, Duong DM, Diner I, Gearing M, Feng Y, Lah JJ, Levey AI, Seyfried NT. Dammer EB, et al. J Proteome Res. 2013 Jul 5;12(7):3193-206. doi: 10.1021/pr400246t. Epub 2013 Jun 17. J Proteome Res. 2013. PMID: 23768213 Free PMC article. - Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins.
Ji Y, Tulin AV. Ji Y, et al. Int J Mol Sci. 2013 Aug 5;14(8):16168-83. doi: 10.3390/ijms140816168. Int J Mol Sci. 2013. PMID: 23921685 Free PMC article. Review. - Changes in the detergent-insoluble brain proteome linked to amyloid and tau in Alzheimer's Disease progression.
Hales CM, Dammer EB, Deng Q, Duong DM, Gearing M, Troncoso JC, Thambisetty M, Lah JJ, Shulman JM, Levey AI, Seyfried NT. Hales CM, et al. Proteomics. 2016 Dec;16(23):3042-3053. doi: 10.1002/pmic.201600057. Proteomics. 2016. PMID: 27718298 Free PMC article. - TDP-43 and ER Stress in Neurodegeneration: Friends or Foes?
de Mena L, Lopez-Scarim J, Rincon-Limas DE. de Mena L, et al. Front Mol Neurosci. 2021 Oct 25;14:772226. doi: 10.3389/fnmol.2021.772226. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34759799 Free PMC article. Review.
References
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
- Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. - PubMed
- Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, et al. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. - PubMed
- Mackenzie IR, Rademakers R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenetics. 2007. - PubMed
- Neumann M, Kwong LK, Truax AC, Vanmassenhove B, Kretzschmar HA, et al. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J Neuropathol Exp Neurol. 2007;66:177–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008169/GM/NIGMS NIH HHS/United States
- F32AG038259/AG/NIA NIH HHS/United States
- F32 AG038259/AG/NIA NIH HHS/United States
- F32 AG032848/AG/NIA NIH HHS/United States
- T32 NS007480/NS/NINDS NIH HHS/United States
- NS-007480/NS/NINDS NIH HHS/United States
- P30 NS055077/NS/NINDS NIH HHS/United States
- F32AG032848-02/AG/NIA NIH HHS/United States
- P30NS055077/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous