Complete genome sequence of Dehalogenimonas lykanthroporepellens type strain (BL-DC-9(T)) and comparison to "Dehalococcoides" strains - PubMed (original) (raw)
. 2012 May 25;6(2):251-64.
doi: 10.4056/sigs.2806097. Epub 2012 May 15.
Jean F Challacombe, Susana F Delano, Lance D Green, Hajnalka Daligault, David Bruce, Chris Detter, Roxanne Tapia, Shunsheng Han, Lynne Goodwin, James Han, Tanja Woyke, Sam Pitluck, Len Pennacchio, Matt Nolan, Miriam Land, Yun-Juan Chang, Nikos C Kyrpides, Galina Ovchinnikova, Loren Hauser, Alla Lapidus, Jun Yan, Kimberly S Bowman, Milton S da Costa, Fred A Rainey, William M Moe
- PMID: 22768368
- PMCID: PMC3387798
- DOI: 10.4056/sigs.2806097
Complete genome sequence of Dehalogenimonas lykanthroporepellens type strain (BL-DC-9(T)) and comparison to "Dehalococcoides" strains
Shivakumara Siddaramappa et al. Stand Genomic Sci. 2012.
Abstract
Dehalogenimonas lykanthroporepellens is the type species of the genus Dehalogenimonas, which belongs to a deeply branching lineage within the phylum Chloroflexi. This strictly anaerobic, mesophilic, non spore-forming, Gram-negative staining bacterium was first isolated from chlorinated solvent contaminated groundwater at a Superfund site located near Baton Rouge, Louisiana, USA. D. lykanthroporepellens was of interest for genome sequencing for two reasons: (a) an unusual ability to couple growth with reductive dechlorination of environmentally important polychlorinated aliphatic alkanes and (b) a phylogenetic position that is distant from previously sequenced bacteria. The 1,686,510 bp circular chromosome of strain BL-DC-9(T) contains 1,720 predicted protein coding genes, 47 tRNA genes, a single large subunit rRNA (23S-5S) locus, and a single, orphan, small subunit rRNA (16S) locus.
Keywords: Chloroflexi; contamination; groundwater; hydrogen utilization; reductive dechlorination; strictly anaerobic.
Figures
Figure 1
Phylogenetic tree showing the position of D. lykanthroporepellens BL-DC-9T relative to other strains within the phylum Chloroflexi. The tree was inferred from 1,214 aligned nucleotide positions of the 16S rRNA gene sequence using the Neighbor-Joining method within the MEGA v4.0.2 package [8]. Truepera radiovictrix RQ-24T is shown as an outgroup and was used to root the tree. Scale bar represents 2 substitutions per 100 nucleotide positions. Numbers at branching points denote support values from 1,000 bootstrap replicates if larger than 70%. Lineages with genome sequencing projects registered in GOLD [9] are shown in blue, published genomes in bold: “_Dehalococcoides_” GT (CP001924), “_Dehalococcoides_” BAV1 (CP000688), “_Dehalococcoides_” CBDB1 (AJ965256), “_Dehalococcoides ethenogenes_” 195 (CP000027), “_Dehalococcoides_” VS (CP001827), Dehalogenimonas lykanthroporepellens BL-DC-9T (CP002084), Caldilinea aerophila STL-6-O1T (AP012337), Anaerolinea thermophila UNI-1T (AP012029), “_Thermobaculum terrenum_” YNP1 (CP001825), Sphaerobacter thermophilus DSM 20745T (CP001823), Thermomicrobium roseum DSM 5159T (CP001275), Herpetosiphon aurantiacus DSM 785T (CP000875), Roseiflexus castenholzii DSM 13941T (CP000804), Chloroflexus aurantiacus J-10-flT (CP000909), and Chloroflexus aggregans MD-66T (CP001337).
Figure 2
Scanning electron micrograph of cells of D. lykanthroporepellens strain BL-DC-9T
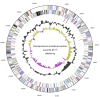
Figure 3
Graphical circular map of the chromosome of D. lykanthroporepellens strain BL-DC-9T. From outside to the center: Genes on the forward strand (color by COG categories), Genes on the reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content (peaks outside the circle indicate above average and peaks inside the circle indicate below average GC content), GC skew [calculated as (G − C) / (G + C); purple indicates values less than 1 and olive indicates values greater than 1]. Numbers outside the map denote nucleotide positions within the chromosome.
Similar articles
- Dehalogenimonas alkenigignens sp. nov., a chlorinated-alkane-dehalogenating bacterium isolated from groundwater.
Bowman KS, Nobre MF, da Costa MS, Rainey FA, Moe WM. Bowman KS, et al. Int J Syst Evol Microbiol. 2013 Apr;63(Pt 4):1492-1498. doi: 10.1099/ijs.0.045054-0. Epub 2012 Aug 10. Int J Syst Evol Microbiol. 2013. PMID: 22888191 - Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater.
Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. Moe WM, et al. Int J Syst Evol Microbiol. 2009 Nov;59(Pt 11):2692-7. doi: 10.1099/ijs.0.011502-0. Epub 2009 Jul 22. Int J Syst Evol Microbiol. 2009. PMID: 19625421 - Dehalogenimonas formicexedens sp. nov., a chlorinated alkane-respiring bacterium isolated from contaminated groundwater.
Key TA, Bowman KS, Lee I, Chun J, Albuquerque L, da Costa MS, Rainey FA, Moe WM. Key TA, et al. Int J Syst Evol Microbiol. 2017 May;67(5):1366-1373. doi: 10.1099/ijsem.0.001819. Epub 2017 Jun 5. Int J Syst Evol Microbiol. 2017. PMID: 28126048 - Genome sequence of the organohalide-respiring Dehalogenimonas alkenigignens type strain (IP3-3(T)).
Key TA, Richmond DP, Bowman KS, Cho YJ, Chun J, da Costa MS, Rainey FA, Moe WM. Key TA, et al. Stand Genomic Sci. 2016 Jun 23;11:44. doi: 10.1186/s40793-016-0165-7. eCollection 2016. Stand Genomic Sci. 2016. PMID: 27340512 Free PMC article. - A case study for microbial biodegradation: anaerobic bacterial reductive dechlorination of polychlorinated biphenyls-from sediment to defined medium.
Bedard DL. Bedard DL. Annu Rev Microbiol. 2008;62:253-70. doi: 10.1146/annurev.micro.62.081307.162733. Annu Rev Microbiol. 2008. PMID: 18729735 Review.
Cited by
- Normalized Quantitative PCR Measurements as Predictors for Ethene Formation at Sites Impacted with Chlorinated Ethenes.
Clark K, Taggart DM, Baldwin BR, Ritalahti KM, Murdoch RW, Hatt JK, Löffler FE. Clark K, et al. Environ Sci Technol. 2018 Nov 20;52(22):13410-13420. doi: 10.1021/acs.est.8b04373. Epub 2018 Nov 8. Environ Sci Technol. 2018. PMID: 30365883 Free PMC article. - Metagenome phylogenetic profiling of microbial community evolution in a tetrachloroethene-contaminated aquifer responding to enhanced reductive dechlorination protocols.
Reiss RA, Guerra P, Makhnin O. Reiss RA, et al. Stand Genomic Sci. 2016 Dec 1;11:88. doi: 10.1186/s40793-016-0209-z. eCollection 2016. Stand Genomic Sci. 2016. PMID: 27980706 Free PMC article. - The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics.
Rupakula A, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J. Rupakula A, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Mar 11;368(1616):20120325. doi: 10.1098/rstb.2012.0325. Print 2013 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23479754 Free PMC article. - Isotopic effects of PCE induced by organohalide-respiring bacteria.
Leitner S, Berger H, Gorfer M, Reichenauer TG, Watzinger A. Leitner S, et al. Environ Sci Pollut Res Int. 2017 Nov;24(32):24803-24815. doi: 10.1007/s11356-017-0075-2. Epub 2017 Sep 15. Environ Sci Pollut Res Int. 2017. PMID: 28913587 - Single cell genomic study of Dehalococcoidetes species from deep-sea sediments of the Peruvian Margin.
Kaster AK, Mayer-Blackwell K, Pasarelli B, Spormann AM. Kaster AK, et al. ISME J. 2014 Sep;8(9):1831-42. doi: 10.1038/ismej.2014.24. Epub 2014 Mar 6. ISME J. 2014. PMID: 24599070 Free PMC article.
References
- Grégoire P, Bohli M, Cayol JL, Joseph M, Guasco S, Dubourg K, Cambar J, Michotey V, Bonin P, Fardeau ML, Ollivier B. Caldilinea tarbellica sp. nov., a filamentous, thermophilic, anaerobic bacterium isolated from a deep hot aquifer in the Aquitaine Basin. Int J Syst Evol Microbiol 2011; 61:1436-1441 10.1099/ijs.0.025676-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases