SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) - PubMed (original) (raw)
. 2012 Oct 11;120(15):3080-8.
doi: 10.1182/blood-2012-01-404863. Epub 2012 Aug 23.
Andreas Roller, Torsten Haferlach, Christiane Eder, Frank Dicker, Vera Grossmann, Alexander Kohlmann, Tamara Alpermann, Kenichi Yoshida, Seishi Ogawa, H Phillip Koeffler, Wolfgang Kern, Claudia Haferlach, Susanne Schnittger
Affiliations
- PMID: 22919025
- PMCID: PMC3580040
- DOI: 10.1182/blood-2012-01-404863
SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML)
Manja Meggendorfer et al. Blood. 2012.
Abstract
We analyzed the mutational hotspot region of SRSF2 (Pro95) in 275 cases with chronic myelomonocytic leukemia (CMML). In addition, ASXL1, CBL, EZH2, JAK2V617F, KRAS, NRAS, RUNX1, and TET2 mutations were investigated in subcohorts. Mutations in SRSF2 (SRSF2mut) were detected in 47% (129 of 275) of all cases. In detail, 120 cases had a missense mutation at Pro95, leading to a change to Pro95His, Pro95Leu, Pro95Arg, Pro95Ala, or Pro95Thr. In 9 cases, 3 new in/del mutations were observed: 7 cases with a 24-bp deletion, 1 case with a 3-bp duplication, and 1 case with a 24-bp duplication. In silico analyses predicted a damaging character for the protein structure of SRSF2 for all mutations. SRSF2mut was correlated with higher age, less pronounced anemia, and normal karyotype. SRSF2mut and EZH2mut were mutually exclusive, but SRSF2mut was associated with TET2mut. In the total cohort, no effect of SRSF2mut on survival was observed. However, in the RUNX1mut subcohort, SRSF2 Pro95His had a favorable effect on overall survival. This comprehensive mutation analysis found that 93% of all patients with CMML carried at least 1 somatic mutation in 9 recurrently mutated genes. In conclusion, these data show the importance of SRSF2mut as new diagnostic marker in CMML.
Figures
Figure 1
Schematic overview of SRSF2 protein organization, mutation type, and mutation frequency. SRSF2 consists of an RRM (AA 14-92; light gray box), a linker region (white box), and an RS-rich domain (AA 117-221; dark gray box). The 4 different mutation types and their mutation localization are indicated in red. The mutation frequency is listed and also shown as black triangles above each mutation type. Each triangle is representing 1 mutated case. From top to bottom, Pro95 missense mutations, p.Pro95_Arg102del;Pro107His, p.Arg86_Gly93dup, and p.Arg94_Pro95insArg alterations are depicted.
Figure 2
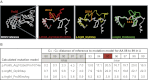
In silico analyses of the structural models of the SRSF2 in/del mutations. The structural changes of the in/del mutations were calculated with the Robetta server algorithm (
). The calculated model for the complete SRSF2wt protein (white structure) is depicted (A), the mutational hotspot Pro95 is marked in red. In addition, the enlargement shows the structure of AA 61-129 of the calculated models: p.[Pro95_Arg102del;Pro107His]; p.Arg86_Gly93dup; and p.Arg94_Pro95insArg. The Cα-Cα distance measurement of the corresponding AA of SRSF2wt to SRSF2mut is shown exemplarily for Pro95. All Å values of the Cα-Cα distance measurements for AA 88-99 are given (B).
Figure 3
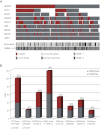
Alignment of gene mutations, karyotype information, and CMML category for 275 patients. (A) Each column represents 1 of the 275 analyzed samples. Analyses of 9 investigated genes, the karyotype, and CMML category-1 or -2 are depicted by colored bars. Red bar indicates mutated gene; dark gray bar, nonmutated gene; white bar, no data available; light-gray bar, normal karyotype; black bar, aberrant karyotype; gray bar, CMML-1; and anthracite bar, CMML-2. (B) Concomitant events of SRSF2 with other mutations are also shown as a bar chart. The gray part represents _SRSF2_wt, the red one _SRSF2_mut within the analyzed subcohorts. _SRSF2_mut frequencies and significances (P values) are denoted; numbers of mutated/analyzed cases of the subcohorts are given in parentheses below the bars..
Figure 4
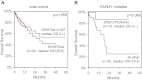
OS by Kaplan-Meier analyses of patients with CMML according to SRSF2 mutations. (A) OS of patients with _SRSF2_mut did not significantly differ from patients with _SRSF2_wt. (B) OS of patients positive for SRSF2 Pro95His compared with patients with all other SRSF2 mutations and _SRSF2_wt (= all other) within the _RUNX1_-mutated subcohort showed a favorable trend. OS is indicated in months and was compared with the 2-sided log-rank test. P values are denoted in each graph, respectively.
Similar articles
- Clinical significance of CSF3R, SRSF2 and SETBP1 mutations in chronic neutrophilic leukemia and chronic myelomonocytic leukemia.
Ouyang Y, Qiao C, Chen Y, Zhang SJ. Ouyang Y, et al. Oncotarget. 2017 Mar 28;8(13):20834-20841. doi: 10.18632/oncotarget.15355. Oncotarget. 2017. PMID: 28209919 Free PMC article. - [SRSF2 mutation in patients with chronic myelomonocytic leukemia].
Yang XC, Zhang SJ, Qiao C, Guo R, Qiu HX, Li JY. Yang XC, et al. Zhonghua Xue Ye Xue Za Zhi. 2013 Dec;34(12):1024-7. doi: 10.3760/cma.j.issn.0253-2727.2013.12.006. Zhonghua Xue Ye Xue Za Zhi. 2013. PMID: 24369158 Chinese. - The detection of SRSF2 mutations in routinely processed bone marrow biopsies is useful in the diagnosis of chronic myelomonocytic leukemia.
Federmann B, Abele M, Rosero Cuesta DS, Vogel W, Boiocchi L, Kanz L, Quintanilla-Martinez L, Orazi A, Bonzheim I, Fend F. Federmann B, et al. Hum Pathol. 2014 Dec;45(12):2471-9. doi: 10.1016/j.humpath.2014.08.014. Epub 2014 Sep 7. Hum Pathol. 2014. PMID: 25305095 - Mutations in chronic myelomonocytic leukemia and their prognostic relevance.
Jian J, Qiao Y, Li Y, Guo Y, Ma H, Liu B. Jian J, et al. Clin Transl Oncol. 2021 Sep;23(9):1731-1742. doi: 10.1007/s12094-021-02585-x. Epub 2021 Apr 16. Clin Transl Oncol. 2021. PMID: 33861431 Review. - Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: a meta-analysis.
Arbab Jafari P, Ayatollahi H, Sadeghi R, Sheikhi M, Asghari A. Arbab Jafari P, et al. Hematology. 2018 Dec;23(10):778-784. doi: 10.1080/10245332.2018.1471794. Epub 2018 May 14. Hematology. 2018. PMID: 29757120 Review.
Cited by
- High-throughput mutational screening adds clinically important information in myelodysplastic syndromes and secondary or therapy-related acute myeloid leukemia.
Karimi M, Nilsson C, Dimitriou M, Jansson M, Matsson H, Unneberg P, Lehmann S, Kere J, Hellström-Lindberg E. Karimi M, et al. Haematologica. 2015 Jun;100(6):e223-5. doi: 10.3324/haematol.2014.118034. Epub 2015 Mar 13. Haematologica. 2015. PMID: 25769547 Free PMC article. No abstract available. - Effective drug treatment identified by in vivo screening in a transplantable patient-derived xenograft model of chronic myelomonocytic leukemia.
Kloos A, Mintzas K, Winckler L, Gabdoulline R, Alwie Y, Jyotsana N, Kattre N, Schottmann R, Scherr M, Gupta C, Adams FF, Schwarzer A, Heckl D, Schambach A, Imren S, Humphries RK, Ganser A, Thol F, Heuser M. Kloos A, et al. Leukemia. 2020 Nov;34(11):2951-2963. doi: 10.1038/s41375-020-0929-3. Epub 2020 Jun 24. Leukemia. 2020. PMID: 32576961 Free PMC article. - SRSF2 mutations in myelodysplasia/myeloproliferative neoplasms.
Aujla A, Linder K, Iragavarapu C, Karass M, Liu D. Aujla A, et al. Biomark Res. 2018 Sep 26;6:29. doi: 10.1186/s40364-018-0142-y. eCollection 2018. Biomark Res. 2018. PMID: 30275952 Free PMC article. Review. - Glycogen synthase kinase-3 and alternative splicing.
Liu X, Klein PS. Liu X, et al. Wiley Interdiscip Rev RNA. 2018 Nov;9(6):e1501. doi: 10.1002/wrna.1501. Epub 2018 Aug 17. Wiley Interdiscip Rev RNA. 2018. PMID: 30118183 Free PMC article. Review. - The mutational landscape of 18 investigated genes clearly separates four subtypes of myelodysplastic/myeloproliferative neoplasms.
Meggendorfer M, Jeromin S, Haferlach C, Kern W, Haferlach T. Meggendorfer M, et al. Haematologica. 2018 May;103(5):e192-e195. doi: 10.3324/haematol.2017.183160. Haematologica. 2018. PMID: 29700173 Free PMC article. No abstract available.
References
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer (IARC); 2008.
- Germing U, Kundgen A, Gattermann N. Risk assessment in chronic myelomonocytic leukemia (CMML). Leuk Lymphoma. 2004;45(7):1311–1318. - PubMed
- Onida F, Kantarjian HM, Smith TL, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99(3):840–849. - PubMed
- Grossmann V, Kohlmann A, Eder C, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(5):877–879. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous