Structural and functional analysis of the NLRP4 pyrin domain - PubMed (original) (raw)
. 2012 Sep 18;51(37):7330-41.
doi: 10.1021/bi3007059. Epub 2012 Sep 6.
Simina Grigoriu, Manuel Hessenberger, Julia Wenger, Sandra Puehringer, Anderson S Pinheiro, Roland N Wagner, Martina Proell, John C Reed, Rebecca Page, Kay Diederichs, Wolfgang Peti
Affiliations
- PMID: 22928810
- PMCID: PMC3445046
- DOI: 10.1021/bi3007059
Structural and functional analysis of the NLRP4 pyrin domain
Clarissa Eibl et al. Biochemistry. 2012.
Abstract
NLRP4 is a member of the nucleotide-binding and leucine-rich repeat receptor (NLR) family of cytosolic receptors and a member of an inflammation signaling cascade. Here, we present the crystal structure of the NLRP4 pyrin domain (PYD) at 2.3 Å resolution. The NLRP4 PYD is a member of the death domain (DD) superfamily and adopts a DD fold consisting of six α-helices tightly packed around a hydrophobic core, with a highly charged surface that is typical of PYDs. Importantly, however, we identified several differences between the NLRP4 PYD crystal structure and other PYD structures that are significant enough to affect NLRP4 function and its interactions with binding partners. Notably, the length of helix α3 and the α2-α3 connecting loop in the NLRP4 PYD are unique among PYDs. The apoptosis-associated speck-like protein containing a CARD (ASC) is an adaptor protein whose interactions with a number of distinct PYDs are believed to be critical for activation of the inflammatory response. Here, we use co-immunoprecipitation, yeast two-hybrid, and nuclear magnetic resonance chemical shift perturbation analysis to demonstrate that, despite being important for activation of the inflammatory response and sharing several similarities with other known ASC-interacting PYDs (i.e., ASC2), NLRP4 does not interact with the adaptor protein ASC. Thus, we propose that the factors governing homotypic PYD interactions are more complex than the currently accepted model, which states that complementary charged surfaces are the main determinants of PYD-PYD interaction specificity.
Figures
Figure 1
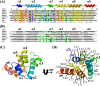
Structural characterization of the NLRP4 PYD. (A) Primary sequence alignment of the NLRP4 PYD and the PYDs of NLRP1, NLRP3, NLRP7, NLRP12, ASC2, and ASC. The six NLRP4 PYD α-helices, illustrated above the sequence, are color-coded from the N-terminus (blue) to the C-terminus (red) and labeled from α1 to α6. Hydrophobic core-forming residues are highlighted in yellow; residues forming the second hydrophobic cluster to stabilize helix α3 are highlighted in green. Positively and negatively charged residues responsible for PYD surface charges are colored blue and red, respectively. Consensus indicates residues that are >90% identical (red) and 50% identical (blue) (! is either Ile or Val; $ is either Leu or Met; # is either Asp or Glu). (B) Comparison of the secondary structural elements of the NLRP4 PYD and the PYDs of NLRP1, NLRP3, NLRP7, NLRP12, ASC2, and ASC (H, helix; L, loop). Helices are labeled from α1 to α6. (C) Cartoon representation of the NLRP4 PYD with the six α-helices colored from the N-terminus (blue) to the C-terminus (red) and labeled from α1 to α6. (D) The NLRP4 PYD (rotated by 90° along the _x_-axis relative to the view in panel (C) is stabilized by an extensive (hydrophobic core residues are labeled and shown as yellow sticks).
Figure 2
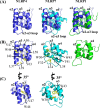
Structural comparison of the NLRP4 PYD with the PYDs of NLRP7 and NLRP1. (A) Superposition of the NLRP4 PYD (blue) on the NLRP7 PYD (cyan) and the NLRP1 PYD (green). (B) Residues that stabilize helix α3 in the NLRP4 PYD and the NLRP7 PYD are labeled and shown as yellow sticks. Corresponding hydrophobic residues are missing in the NLRP1 PYD. (C) Trp44 in the NLRP4 PYD is structurally conserved in the NLRP7 PYD.
Figure 3
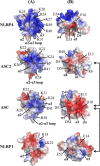
Electrostatic surface potentials of the NLRP4 PYD compared to ASC2 and the PYDs of ASC and NLRP1. The electrostatic potentials (ESPs) of the two interaction surfaces formed by helices α2 and α3 and helices α1 and α4 are mapped on the solvent-accessible surface of the identically oriented PYDs from NLRP4, NLRP1, ASC, and ASC2. The color scale varies from red (negative ESP) to blue (positive ESP). Residues responsible for these charges are labeled on each structure. Arrows indicate previously experimentally detected direct homtoypic PYD interactions.
Figure 4
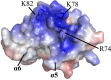
NLRP4 PYD showing an additional positive patch. The electrostatic potentials of helices α5 and α6 are mapped onto the solvent-accessible surface of the NLRP4 PYD. Residues Arg74, Lys78, and Lys82, form this positively charged surface.
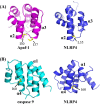
Figure 5
Comparison of the NLRP4 PYD with the Apaf-1 CARD and the caspase 9 CARD. Residues (yellow sticks) that mediate a hydrophobic interaction between the Apaf-1 CARD (magenta) and the caspase 9 CARD (cyan) are also surface-exposed in the NLRP4 PYD (blue).
Similar articles
- Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly.
Jin T, Perry A, Smith P, Jiang J, Xiao TS. Jin T, et al. J Biol Chem. 2013 May 10;288(19):13225-35. doi: 10.1074/jbc.M113.468033. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530044 Free PMC article. - Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity.
Pinheiro AS, Proell M, Eibl C, Page R, Schwarzenbacher R, Peti W. Pinheiro AS, et al. J Biol Chem. 2010 Aug 27;285(35):27402-27410. doi: 10.1074/jbc.M110.113191. Epub 2010 Jun 11. J Biol Chem. 2010. PMID: 20547486 Free PMC article. - Structure and dynamics of ASC2, a pyrin domain-only protein that regulates inflammatory signaling.
Natarajan A, Ghose R, Hill JM. Natarajan A, et al. J Biol Chem. 2006 Oct 20;281(42):31863-75. doi: 10.1074/jbc.M605458200. Epub 2006 Aug 10. J Biol Chem. 2006. PMID: 16905547 - Inhibiting the inflammasome: one domain at a time.
Dorfleutner A, Chu L, Stehlik C. Dorfleutner A, et al. Immunol Rev. 2015 May;265(1):205-16. doi: 10.1111/imr.12290. Immunol Rev. 2015. PMID: 25879295 Free PMC article. Review. - Fire and death: the pyrin domain joins the death-domain superfamily.
Kohl A, Grütter MG. Kohl A, et al. C R Biol. 2004 Dec;327(12):1077-86. doi: 10.1016/j.crvi.2004.08.006. C R Biol. 2004. PMID: 15656350 Review.
Cited by
- Structure, interactions and self-assembly of ASC-dependent inflammasomes.
de Alba E. de Alba E. Arch Biochem Biophys. 2019 Jul 30;670:15-31. doi: 10.1016/j.abb.2019.05.023. Epub 2019 May 30. Arch Biochem Biophys. 2019. PMID: 31152698 Free PMC article. Review. - Role of NLRs in the Regulation of Type I Interferon Signaling, Host Defense and Tolerance to Inflammation.
Kienes I, Weidl T, Mirza N, Chamaillard M, Kufer TA. Kienes I, et al. Int J Mol Sci. 2021 Jan 28;22(3):1301. doi: 10.3390/ijms22031301. Int J Mol Sci. 2021. PMID: 33525590 Free PMC article. Review. - Role of Microgliosis and NLRP3 Inflammasome in Parkinson's Disease Pathogenesis and Therapy.
de Araújo FM, Cuenca-Bermejo L, Fernández-Villalba E, Costa SL, Silva VDA, Herrero MT. de Araújo FM, et al. Cell Mol Neurobiol. 2022 Jul;42(5):1283-1300. doi: 10.1007/s10571-020-01027-6. Epub 2021 Jan 2. Cell Mol Neurobiol. 2022. PMID: 33387119 Review. - NOD-Like Receptors in Lung Diseases.
Chaput C, Sander LE, Suttorp N, Opitz B. Chaput C, et al. Front Immunol. 2013 Nov 21;4:393. doi: 10.3389/fimmu.2013.00393. Front Immunol. 2013. PMID: 24312100 Free PMC article. Review. - An updated view on the structure and function of PYRIN domains.
Chu LH, Gangopadhyay A, Dorfleutner A, Stehlik C. Chu LH, et al. Apoptosis. 2015 Feb;20(2):157-73. doi: 10.1007/s10495-014-1065-1. Apoptosis. 2015. PMID: 25451010 Free PMC article.
References
- Kersse K.; Bertrand M. J.; Lamkanfi M.; Vandenabeele P. (2011) NOD-like receptors and the innate immune system: Coping with danger, damage and death. Cytokine Growth Factor Rev. 22, 257–276. - PubMed
- Martinon F.; Tschopp J. (2005) NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 26, 447–454. - PubMed
- Fritz J. H.; Ferrero R. L.; Philpott D. J.; Girardin S. E. (2006) Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 7, 1250–1257. - PubMed
- Martinon F.; Burns K.; Tschopp J. (2002) The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426. - PubMed
- Petrilli V.; Dostert C.; Muruve D. A.; Tschopp J. (2007) The inflammasome: A danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 19, 615–622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- J 3154/FWF_/Austrian Science Fund FWF/Austria
- J 3173/FWF_/Austrian Science Fund FWF/Austria
- T32 GM007601/GM/NIGMS NIH HHS/United States
- R01-AI-56324/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous