Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia - PubMed (original) (raw)
Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia
Max Jan et al. Sci Transl Med. 2012.
Abstract
Given that most bone marrow cells are short-lived, the accumulation of multiple leukemogenic mutations in a single clonal lineage has been difficult to explain. We propose that serial acquisition of mutations occurs in self-renewing hematopoietic stem cells (HSCs). We investigated this model through genomic analysis of HSCs from six patients with de novo acute myeloid leukemia (AML). Using exome sequencing, we identified mutations present in individual AML patients harboring the FLT3-ITD (internal tandem duplication) mutation. We then screened the residual HSCs and detected some of these mutations including mutations in the NPM1, TET2, and SMC1A genes. Finally, through single-cell analysis, we determined that a clonal progression of multiple mutations occurred in the HSCs of some AML patients. These preleukemic HSCs suggest the clonal evolution of AML genomes from founder mutations, revealing a potential mechanism contributing to relapse. Such preleukemic HSCs may constitute a cellular reservoir that should be targeted therapeutically for more durable remissions.
Conflict of interest statement
COMPETING INTERESTS.
The authors declare no competing financial interests.
Figures
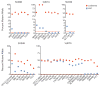
Figure 1. Prospective separation of residual HSCs from leukemia cells
(A) Flow cytometry analysis of samples from AML cases SU008 and SU048 indicating lineage negative cells (left panels) and Lin−CD34+CD38− subsets (right panels) further analyzed for expression of CD99 and/or TIM3. Leukemia cells (red gate) and putative residual HSCs (blue gate) were each purified via two rounds of FACS (Supplemental Figure 1). (B) Lin−CD34+CD38−CD99− cells (6000 cells) isolated from case SU008 or Lin−CD34+CD38−TIM3−CD99− cells (450 cells) isolated from case SU048 were transplanted into newborn NSG mice. 12 weeks later, bone marrow engraftment was analyzed by flow cytometry for the presence of human hematopoietic cells (hCD45+) (left panel), further subdivided into lymphoid (CD19+) and myeloid (CD33+) subsets (right panel). (C) The indicated cells were further analyzed for the presence of the _FLT3_-ITD mutation by PCR. Leukemia cells and HSCs from each case corresponded to the red and blue gates in panel A, respectively, and the analysis also included bone marrow from mice engrafted with the residual HSCs. (D) Experimental scheme for identification of pre-leukemic mutations and clonal evolution in de novo AML.
Figure 2. Targeted sequencing identifies pre-leukemic HSCs
FACS-purified leukemia cells and residual HSCs from AML cases SU008, SU014, SU030, SU048, and SU070 were analyzed by targeted deep sequencing for the presence of patient-matched somatic mutations in genes with detectable mRNA expression identified in leukemia cells by exome and transcriptome sequencing. The percentage of mutant allele reads is indicated in each case. The dashed red line indicates the threshold of 1% for variant allele detection as determined by sequencing of defined mixtures of normal and leukemic DNA (fig. S6). Details of sequencing reads are presented in table S2.
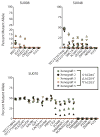
Figure 3. Functional HSCs contain pre-leukemic mutations
FACS-purified hCD45+, hCD45+CD19+, and/or hCD45+CD33+ cells isolated from the bone marrow of mice engrafted with residual HSCs from AML cases SU008 (n=4), SU048 (n=5), and SU070 (n=3) were analyzed by targeted deep sequencing for patient-matched somatic mutations. The percentage of mutant allele reads is indicated in each case. The dashed red line indicates the threshold of 1% for variant allele detection as determined by sequencing of defined mixtures of normal and leukemic DNA (fig. S6).
Figure 4. Single cell analysis identifies sequential mutation acquisition in pre-leukemic HSCs
(A) The genotype of 546 myeloid colonies derived from clone sorted single residual HSCs from case SU008 were determined by multiplexed custom Taqman SNP assays for mutations identified by exome analysis to be present in each population. Selected assays are shown with the full data presented in fig. S8. Each colony is represented by a single dot in graphs for each mutation tested; colonies are colored according to genotype (see sure key). (B) 3-D plots illustrate the genotype of each colony. In both panels, the yellow dot indicates the genotype of patient T cells, and the red dot indicates the genotype of patient leukemia cells. (C) Shown is a model for the proposed clonal evolution of AML in patient SU008.
Figure 5. Single cell analysis identifies sequential driver mutation acquisition in pre-leukemic HSCs
(A, D) The genotype of 81 myeloid colonies derived from clone sorted single residual HSCs from AML case SU048 (A) or 189 colonies derived from single HSCs from AML case SU070 (D) were determined by multiplexed custom Taqman SNP assays for mutations identified by exome analysis to be present in each population. Selected assays are shown with the full data presented in fig. S10 (SU048) and fig. S11 (SU070). Each colony is represented by a single dot in graphs for each mutation tested; colonies are colored according to genotype (see figure key). (B, E) 3-D plots illustrating the genotype of each colony from SU048 (B) and SU070 (E). In both panels, the yellow dot indicates the genotype of patient T cells, and the red dot indicates the genotype of patient leukemia cells. (C, F) Models for the proposed clonal evolution of SU048 (C) and SU070 (F) are presented.
Similar articles
- Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission.
Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Corces-Zimmerman MR, et al. Proc Natl Acad Sci U S A. 2014 Feb 18;111(7):2548-53. doi: 10.1073/pnas.1324297111. Epub 2014 Feb 3. Proc Natl Acad Sci U S A. 2014. PMID: 24550281 Free PMC article. - Clonal evolution of pre-leukemic hematopoietic stem cells precedes human acute myeloid leukemia.
Majeti R. Majeti R. Best Pract Res Clin Haematol. 2014 Sep-Dec;27(3-4):229-34. doi: 10.1016/j.beha.2014.10.003. Epub 2014 Oct 15. Best Pract Res Clin Haematol. 2014. PMID: 25455271 Free PMC article. Review. - Bone Marrow Clonogenic Myeloid Progenitors from _NPM1_-Mutated AML Patients Do Not Harbor the NPM1 Mutation: Implication for the Cell-Of-Origin of NPM1+ AML.
Guardia RD, González-Silva L, López-Millán B, Rodríguez-Sevilla JJ, Baroni ML, Bueno C, Anguita E, Vives S, Palomo L, Lapillonne H, Varela I, Menendez P. Guardia RD, et al. Genes (Basel). 2020 Jan 9;11(1):73. doi: 10.3390/genes11010073. Genes (Basel). 2020. PMID: 31936647 Free PMC article. - Genomic analysis of cellular hierarchy in acute myeloid leukemia using ultrasensitive LC-FACSeq.
Saygin C, Hu E, Zhang P, Sher S, Lozanski A, Doong TJ, Nicolet D, Orwick S, Labanowska J, Skinner JN, Cempre C, Kauffman T, Goettl VM, Heerema NA, Abruzzo L, Miller C, Lapalombella R, Behbehani G, Mims AS, Larkin K, Grieselhuber N, Walker A, Bhatnagar B, Bloomfield CD, Byrd JC, Lozanski G, Blachly JS. Saygin C, et al. Leukemia. 2021 Dec;35(12):3406-3420. doi: 10.1038/s41375-021-01295-1. Epub 2021 May 21. Leukemia. 2021. PMID: 34021247 Free PMC article. - Cellular carcinogenesis in preleukemic conditions:drivers and defenses.
Ueda K, Ikeda K. Ueda K, et al. Fukushima J Med Sci. 2024 Jan 27;70(1):11-24. doi: 10.5387/fms.2023-17. Epub 2023 Nov 11. Fukushima J Med Sci. 2024. PMID: 37952978 Free PMC article. Review.
Cited by
- Multi-omic analysis of longitudinal acute myeloid leukemia patient samples reveals potential prognostic markers linked to disease progression.
Ahmed N, Cavattoni I, Villiers W, Cugno C, Deola S, Mifsud B. Ahmed N, et al. Front Genet. 2024 Sep 27;15:1442539. doi: 10.3389/fgene.2024.1442539. eCollection 2024. Front Genet. 2024. PMID: 39399221 Free PMC article. - Defining heritability, plasticity, and transition dynamics of cellular phenotypes in somatic evolution.
Schiffman JS, D'Avino AR, Prieto T, Pang Y, Fan Y, Rajagopalan S, Potenski C, Hara T, Suvà ML, Gawad C, Landau DA. Schiffman JS, et al. Nat Genet. 2024 Oct;56(10):2174-2184. doi: 10.1038/s41588-024-01920-6. Epub 2024 Sep 24. Nat Genet. 2024. PMID: 39317739 - Early drivers of clonal hematopoiesis shape the evolutionary trajectories of de novo acute myeloid leukemia.
Chow RD, Velu P, Deihimi S, Belman J, Youn A, Shah N, Luger SM, Carroll MP, Morrissette J, Bowman RL. Chow RD, et al. medRxiv [Preprint]. 2024 Sep 1:2024.08.31.24312756. doi: 10.1101/2024.08.31.24312756. medRxiv. 2024. PMID: 39252918 Free PMC article. Preprint. - Mutational cooperativity of RUNX1::RUNX1T1 isoform 9a and oncogenic NRAS in zebrafish myeloid leukaemia.
Lints R, Walker CA, Delfi O, Prouse M, PohLui De Silva M, Bohlander SK, Wood AC. Lints R, et al. Biol Open. 2024 Sep 15;13(9):bio060523. doi: 10.1242/bio.060523. Epub 2024 Aug 30. Biol Open. 2024. PMID: 39177514 Free PMC article. - The profile and prognostic significance of bone marrow T-cell differentiation subsets in adult AML at diagnosis.
Sun K, Shi ZY, Wang YZ, Xie DH, Liu YR, Jiang Q, Jiang H, Huang XJ, Qin YZ. Sun K, et al. Front Immunol. 2024 Jul 19;15:1418792. doi: 10.3389/fimmu.2024.1418792. eCollection 2024. Front Immunol. 2024. PMID: 39100667 Free PMC article.
References
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. - PubMed
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. - PubMed
- Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Dohner H. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. - PubMed
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A, Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC, Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. - PMC - PubMed
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- U01 HL099995/HL/NHLBI NIH HHS/United States
- U01 HL099999/HL/NHLBI NIH HHS/United States
- R01 CA086017/CA/NCI NIH HHS/United States
- R01 CA086065/CA/NCI NIH HHS/United States
- G1000729/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous