Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans - PubMed (original) (raw)
Review
Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans
Karen Adelman et al. Nat Rev Genet. 2012 Oct.
Abstract
Recent years have witnessed a sea change in our understanding of transcription regulation: whereas traditional models focused solely on the events that brought RNA polymerase II (Pol II) to a gene promoter to initiate RNA synthesis, emerging evidence points to the pausing of Pol II during early elongation as a widespread regulatory mechanism in higher eukaryotes. Current data indicate that pausing is particularly enriched at genes in signal-responsive pathways. Here the evidence for pausing of Pol II from recent high-throughput studies will be discussed, as well as the potential interconnected functions of promoter-proximally paused Pol II.
Figures
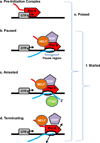
Figure 1. Defining the terms used to describe promoter-associated Pol II complexes
The promoter region is depicted, with the transcription start site (TSS) labelled with an arrow. Pol II is illustrated as a red rocket. The general transcription factors (GTFs, grey oval) are shown centred at the TSS (arrow). The pause-inducing factors NELF (orange oval), DSIF (purple pentagon) and transcript cleavage factor TFIIS (green circle) are shown. The nascent RNA transcript is shown in blue and a bracket indicates the pausing region, usually 20–60 nucleotides downstream from the TSS.
- Pre-Initiation Complex: an entry form of Pol II in complex with general transcription factors, where the polymerase is bound to the promoter DNA but has not yet initiated RNA synthesis.
- Paused: an early elongation complex that has transiently halted RNA synthesis. Paused polymerase is fully competent to resume elongation, remaining stably engaged and associated with the nascent RNA. The 3’-end of the RNA may have ‘frayed’ slightly from the Pol II active site in a manner that would slow further RNA synthesis, but the RNA is properly aligned with the active site. Two protein complexes, DSIF and NELF reduce the rate of elongation and facilitate the establishment of the stably paused state.
- Arrested: a stably engaged elongation complex wherein the polymerase has backtracked along the DNA template, such that the RNA 3’-end is displaced from the active site. Re-start of an arrested complex usually requires the transcript cleavage factor TFIIS, which induces Pol II to cleave the nascent RNA at the active site, creating a new 3’-end that is properly aligned with the Pol II active site and releasing a short (2–9 nt) 3’ RNA.
- Terminating: an unstable elongation complex that is in the process of dissociating from the DNA template and releasing the nascent RNA. The released Pol II could have the potential to rapidly re-initiate transcription and “re-cycle” at the promoter.
- Poised: very generic term that simply indicates that Pol II is located near the TSS, but does not specify anything about its transcriptional status. Can include any of the above complexes (a–d).
- Stalled: term indicating that Pol II is engaged in transcription, but that makes no assumptions about its ability to resume synthesis. This term includes paused, arrested and terminating complexes (b–d, above).
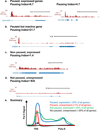
Figure 2. Patterns of Pol II distribution across gene regions
Pol II ChIP-seq signal is shown at genes that exemplify a variety of Pol II distributions that are observed across metazoan genomes. Example genes are taken from Drosophila S2 cells, and the pausing index of each gene is indicated.
- Two paused, but active genes with differing pausing indices
- A paused but inactive gene
- A Non-paused gene that is expressed
- A Non-paused, unexpressed gene. We note that a pausing index cannot reliably be calculated for genes that lack significant Pol II promoter signal.
- Shown are profiles of Pol II signal that exemplify the four major groups of genes. We note that the “paused, unexpressed” group is significantly under-represented in vivo, suggesting that most paused genes display some basal RNA synthesis. Approximate percentages of genes that fall into each category are given, , .
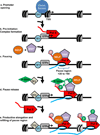
Figure 3. Establishment and release of paused Pol II
The promoter region is shown with the transcription start site (TSS) labelled with an arrow. Nucleosomes are depicted in grey and Pol II is illustrated as a red rocket. The nascent RNA transcript is shown in blue and a bracket indicates the region wherein pausing takes place. Factors involved in the establishment or release of paused Pol II, such as DSIF (purple pentagon), NELF (orange oval) and P-TEFb (green diamond) are indicated.
- Promoter opening often involves the binding of a sequence specific transcription factor (shown here as TF1, light blue circle) that brings in chromatin remodelers (blue oval) to remove nucleosomes from around the TSS and render the promoter accessible for recruitment of the transcription machinery.
- Pre-Initiation Complex (PIC) formation involves the recruitment of a set of general transcription factors (GTFs, grey oval) and Pol II which is also facilitated by the binding of specific transcription factors (also shown as TF1 for simplicity). This step precedes the initiation of RNA synthesis.
- Pol II pausing occurs shortly after transcription initiation and involves the association of pausing factors DSIF and NELF. The paused Pol II is phosphorylated on its CTD (shown in pink). The region wherein pausing takes place is shown by a bracket.
- Pause release is triggered by the recruitment of the P-TEFb kinase (green diamond), either directly or indirectly by a transcription factor (shown here as TF2, tan diamond). P-TEFb kinase phosphorylates the DSIF/NELF complex to release paused Pol II and also targets the CTD (shown in green). Phosphorylation of DSIF/NELF dissociates NELF from the elongation complex and transforms DSIF into a positive elongation factor that associates with Pol II throughout the gene.
- In the presence of both TF1 and TF2, escape of the paused Pol II into productive elongation is followed rapidly by entry of another Pol II into the pause site, allowing for efficient RNA production. When the gene is activated, some nucleosome disruption is likely, as depicted by the lighter colouring of the downstream nucleosome.
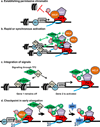
Figure 4. Illustrations of the main hypotheses for the functions of Pol II pausing
The promoter region is depicted, with the transcription start site (TSS) labelled with an arrow. Nucleosomes are shown in grey and Pol II is illustrated as a red rocket. The general transcription factors (GTFs) are shown centred at the TSS (grey oval). The pause-inducing factors NELF (orange oval) and DSIF (purple pentagon) are shown. The nascent RNA transcript is shown in blue.
- Establishing permissive chromatin: in the absence of paused Pol II, nucleosomes occlude the promoter region and inhibit gene expression, as shown in the left hand panel. Once, these nucleosomes are remodelled, paused Pol II helps to maintain the nucleosome-deprived structure by blocking nucleosome assembly over promoter sequences (depicted at right). Pausing thus would keep the promoter region accessible for activator and transcription factor binding.
- Rapid or synchronous gene activation: at a gene with paused Pol II, gene activation could proceed simply through recruitment of P-TEFb, thereby triggering the rapid release of paused Pol II into productive elongation. If a number of genes harbouring paused Pol II were all activated by the same signal and associated transcription factor (shown as TF2), then these genes could activated simultaneously and in a rapid fashion.
- Integrating multiple regulatory signals: Pausing represents a separate step in the transcription cycle for factors to act, and allows for combinatorial control between transcription factors that recruit the transcription machinery (TF1) and those that trigger pause release (TF2), where both would be necessary for gene activation. In this example, signalling through TF2 such that it binds DNA and recruits P-TEFb would not lead to activation of a gene that was not paused (i.e. lacks TF1), but would stimulate synthesis from Gene 2 that was loaded with paused polymerase.
- Checkpoint in early elongation: on the left, arrows depict interactions between the Capping Enzyme Complex (CEC) and DSIF/NELF as well as the Ser5-P modification of the CTD of Pol II, which is thought to stimulate capping activity. The hat represents the 5’ RNA cap. Center, P-TEFb dependent phosphorylation events release paused Pol II, and create a platform for binding of RNA processing factors (RPF) on the Ser2-P CTD of Pol II, as shown at right.
Similar articles
- Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans.
Dollinger R, Gilmour DS. Dollinger R, et al. J Mol Biol. 2021 Jul 9;433(14):166897. doi: 10.1016/j.jmb.2021.166897. Epub 2021 Feb 25. J Mol Biol. 2021. PMID: 33640324 Free PMC article. Review. - Promoter proximal pausing on genes in metazoans.
Gilmour DS. Gilmour DS. Chromosoma. 2009 Feb;118(1):1-10. doi: 10.1007/s00412-008-0182-4. Epub 2008 Oct 2. Chromosoma. 2009. PMID: 18830703 Review. - Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila.
Ivaldi MS, Karam CS, Corces VG. Ivaldi MS, et al. Genes Dev. 2007 Nov 1;21(21):2818-31. doi: 10.1101/gad.1604007. Epub 2007 Oct 17. Genes Dev. 2007. PMID: 17942706 Free PMC article. - The Hox proteins Ubx and AbdA collaborate with the transcription pausing factor M1BP to regulate gene transcription.
Zouaz A, Auradkar A, Delfini MC, Macchi M, Barthez M, Ela Akoa S, Bastianelli L, Xie G, Deng WM, Levine SS, Graba Y, Saurin AJ. Zouaz A, et al. EMBO J. 2017 Oct 2;36(19):2887-2906. doi: 10.15252/embj.201695751. Epub 2017 Sep 4. EMBO J. 2017. PMID: 28871058 Free PMC article. - Promoter-proximal pausing on the hsp70 promoter in Drosophila melanogaster depends on the upstream regulator.
Tang H, Liu Y, Madabusi L, Gilmour DS. Tang H, et al. Mol Cell Biol. 2000 Apr;20(7):2569-80. doi: 10.1128/MCB.20.7.2569-2580.2000. Mol Cell Biol. 2000. PMID: 10713179 Free PMC article.
Cited by
- Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq).
Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. Mahat DB, et al. Nat Protoc. 2016 Aug;11(8):1455-76. doi: 10.1038/nprot.2016.086. Epub 2016 Jul 21. Nat Protoc. 2016. PMID: 27442863 Free PMC article. - Cell-type-specific loops linked to RNA polymerase II elongation in human neural differentiation.
Titus KR, Simandi Z, Chandrashekar H, Paquet D, Phillips-Cremins JE. Titus KR, et al. Cell Genom. 2024 Aug 14;4(8):100606. doi: 10.1016/j.xgen.2024.100606. Epub 2024 Jul 10. Cell Genom. 2024. PMID: 38991604 Free PMC article. - Live-cell imaging of RNA Pol II and elongation factors distinguishes competing mechanisms of transcription regulation.
Versluis P, Graham TGW, Eng V, Ebenezer J, Darzacq X, Zipfel WR, Lis JT. Versluis P, et al. Mol Cell. 2024 Aug 8;84(15):2856-2869.e9. doi: 10.1016/j.molcel.2024.07.009. Mol Cell. 2024. PMID: 39121843 - Exploring the interplay between PARP1 and circRNA biogenesis and function.
Dhahri H, Fondufe-Mittendorf YN. Dhahri H, et al. Wiley Interdiscip Rev RNA. 2023 Nov 13:e1823. doi: 10.1002/wrna.1823. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37957925 Review. - RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription.
Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, Piotrowski J, Thomas CJ, Singer DS, Pugh BF, Pommier Y, Przytycka TM, Kouzine F, Lewis BA, Zhao K, Levens D. Baranello L, et al. Cell. 2016 Apr 7;165(2):357-71. doi: 10.1016/j.cell.2016.02.036. Cell. 2016. PMID: 27058666 Free PMC article.
References
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. - PubMed
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848.. Global run-on sequencing (GRO-seq) maps the position, amount, and orientation of transcriptionally engaged RNA polymerases genome-wide, and shows peaks of promoter-proximal polymerase residing on ~30% of human genes.
- Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551.. Global analyses of Pol II pausing and nucleosome occupancy reveal that Pol II and nucleosomes compete for promoter occupancy to co-ordinately regulate gene expression.
- Lee C, et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300.. Comprehensive analysis of promoter-associated Pol II in Drosophila using ChIP-chip and permanganate demonstrates that NELF-mediated pausing of Pol II is common in Drosophila.
Publication types
MeSH terms
Substances
Grants and funding
- ZIA ES101987-07/Intramural NIH HHS/United States
- GM25232/GM/NIGMS NIH HHS/United States
- R37 GM025232/GM/NIGMS NIH HHS/United States
- Z01 ES101987/Intramural NIH HHS/United States
- R01 GM025232/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources