Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate - PubMed (original) (raw)
Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate
Byung Hak Ha et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The type II p21-activated kinases (PAKs) are key effectors of RHO-family GTPases involved in cell motility, survival, and proliferation. Using a structure-guided approach, we discovered that type II PAKs are regulated by an N-terminal autoinhibitory pseudosubstrate motif centered on a critical proline residue, and that this regulation occurs independently of activation loop phosphorylation. We determined six X-ray crystal structures of either full-length PAK4 or its catalytic domain, that demonstrate the molecular basis for pseudosubstrate binding to the active state with phosphorylated activation loop. We show that full-length PAK4 is constitutively autoinhibited, but mutation of the pseudosubstrate releases this inhibition and causes increased phosphorylation of the apoptotic regulation protein Bcl-2/Bcl-X(L) antagonist causing cell death and cellular morphological changes. We also find that PAK6 is regulated by the pseudosubstrate region, indicating a common type II PAK autoregulatory mechanism. Finally, we find Src SH3, but not β-PIX SH3, can activate PAK4. We provide a unique understanding for type II PAK regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
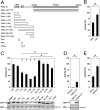
PAK schematic and kinase assays for type II PAKs. (A) Schematic diagram for type I and type II PAK family members. The type I PAKs contain an N-terminal GBD that overlaps with an AID (GBD:AID), and a C-terminal kinase domain. The type II PAKs contain an N-terminal GBD and a C-terminal kinase domain. All PAKs contain proline-rich patches between the N-terminal GBD and C-terminal kinase domains. (B) Kinase assay for PAK4-cat and PAK4-FL. Purified PAK4 catalytic domain PAK4-cat135–426 (PAK4-cat and PAK4-FL were assayed for activity toward MBP with [γ-33P]ATP. Reactions were subjected to SDS/PAGE, exposed to phosphor storage screen, scanned, and MBP phosphorylation quantified by optical densitometry (Lower, MBP-P). MBP loading is shown (Lower, MBP, Coomassie). Activities are shown as a percentage of PAK4-cat activity (100%). PAK4-FL displays ∼100-fold less activity than PAK4-cat. *P < 0.01, t test. (C) GTPases do not increase PAK4-FL catalytic activity. PAK4-FL kinase activity toward MBP was assayed in the presence of either purified CDC42 or RAC1 with GMP-PNP and Mg2+. MBP and GTPase loading are shown. Small GTPases do not impact kinase activity of PAK4-FL toward MBP.
Fig. 2.
Structures of PAK4-cat and PAK4-FL in the _P_3 crystal form. (A) Overall structure of the PAK4 kinase domain. PAK4 kinase domain shown in ribbon format with N-lobe (light gray) and C-lobe (dark gray) indicated. Bound AMP-PNP shown in stick format. N and C termini are indicated. Region shown in B and C is indicted by a dashed box. (B and C) The PAK4-FL _P_3 crystal contains a significant region of positive difference electron density in the peptide substrate binding site; this is not observed in the PAK4-cat structure. Two contour levels (2σ and 3σ in dark and light green, respectively) are shown for the unbiased _F_obs-_F_calc map. (D) Sequence alignment for the N terminus of human type II PAKs (UniProt accession nos: PAK4, O96013; PAK5, Q9P286; PAK6, Q9NQU5). The four PAK4 isoforms are shown, which are identical between the N terminus and residue K68. (*) indicates identical; (:), highly conserved; (.), semiconserved. The PAK4 GBD (CRIB) domain as defined by Abo et al. (5) is shaded yellow. QKF peptide, green; RPK peptide, red; and 23-mer peptide, blue.
Fig. 3.
PAK4 kinase assays. (A) Schematic of constructs used for PAK4 kinase activity assays. Residues mutated are indicated on PAK4-FL as black lines with two filled circles (••) indicating R49PKP to A49AAA or a single filled circle (•) indicating I59T to A59A. (B) Purified PAK4 catalytic domain PAK4-cat135–426 (PAK4-cat) was assayed for activity toward MBP in the presence of RPK or QKF peptides. Radiolabel incorporation into MBP was determined by optical densitometry following SDS/PAGE and exposure to a phosphor storage screen. Activities are shown as a percentage of the activity of PAK4-cat135–426. For all, kinase assays show SEM for more than three experiments. RPK peptide significantly inhibits kinase activity. *P < 0.01, t test. (C) PAK4-cat135–426 activity in the presence of N-terminal constructs of PAK4. Purified MalBP fusion constructs PAK41–30, PAK41–45, PAK41–70, PAK41–90, PAK41–130, PAK431–130, PAK480–130, and PAK4110–130 were added and compared with addition of MalBP alone (MalBP). PAK41–70, PAK41–90, PAK41–130, and PAK431–130 inhibit kinase activity of PAK4-cat135–426. MBP phosphorylation (Lower, MBP-P) and loading are shown (Lower, MBP, Coomassie). Mutation of R49PKP to A49AAA in either PAK41–70 or PAK41–130 results in a loss of inhibition of kinase activity. Activities are shown as a percentage of the activity of PAK4-cat135–426 with negative control MalBP added. *P < 0.01, t test; +P < 0.01 by t test compared with PAK4-cat135–426 with MalBP alone. (D) Mutation of PAK4-FL restores kinase activity. Purified PAK4-FL containing point mutations R49PKP to A49AAA (PAK4-FL4A) show a significant increase kinase activity with respect to MBP. Activities are shown as a percentage of PAK4-cat135–426 activity. (E) Effect of 23-mer peptide on PAK4-cat135–426 kinase activity. The 23-mer peptide inhibits kinase activity better than the RPK peptide (B). Mutation of R49PKP to A49AAA and I59T to A59A in this peptide (23-mer6A) results in a loss of kinase inhibition. Activities are shown as a percentage of the activity of PAK4-cat135–426.
Fig. 4.
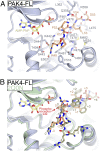
Mode of binding for the type II PAK autoinhibitory region. (A) Structural details of pseudosubstrate binding to PAK4 catalytic domain. Structure of PAK4-FL is shown with residues discussed in the text shown in stick format and labeled. Pseudosubstrate is colored orange. Hydrogen bonds are colored green. (B) Comparison of pseudosubstrate binding to a consensus substrate peptide. Crystal structure of PAK4 catalytic domain bound to a consensus substrate sequence (PDB ID code 2Q0N) is shown in green. Labels for substrate and pseudosubstrate indicate number of residues distal from the phosphoacceptor site (labeled and denoted 0).
Fig. 5.
Cellular effects of loss of PAK4 pseudosubstrate inhibition and PAK6 pseudosubstrate inhibition. (A) Morphological changes upon transient expression of PAK4-FL or PAK4-FL4A. NIH 3T3 cells transiently transfected with GFP-PAK4-FL4A (PAK4-FL4A) exhibit cell rounding that is not observed with GFP-PAK4-FL (PAK4-FL). (B and C) Reduced actin containing stress fibers in COS-7 cells expressing activated PAK4. Transiently expressed GFP-tagged GFP-PAK4-FL (B) or GFP-PAK4-FL4A (C) were fixed with paraformaldehyde, permeabilized, then stained with rhodamine-conjugated phalloidin and imaged with a spinning-disk confocal-based inverted Olympus microscope. The overall size and extent of stress fiber organization (Insets and arrows) was decreased in cells expressing the PAK4-FL4A mutant compared with cells expressing PAK4-FL. Insets and arrows show reduced stress fibers organization and increase in actin containing membrane ruffles in cells expressing the PAK4-FL4A mutant. Transfection of cells with GFP-PAK4 constructs is validated in
Fig. S4
. (Scale bars: 10 μm.) (D) Phosphorylation of the PAK4 substrate BAD. Transient overexpression of GFP-PAK4-FL (WT), GFP-PAK4-FL4A (4A), or GFP-PAK4-cat135–426 shows that mutation of the PAK4 pseudosubstrate elevates BAD Ser112 phosphorylation to a level equivalent to that induced by the kinase domain alone (cat). (E) Purified PAK6 catalytic domain (PAK6-cat) was assayed for activity toward MBP in the presence of the 23-mer and 23-mer6A peptides. MBP was loaded onto SDS/PAGE, exposed to phosphor storage screen, scanned, and quantified by optical densitometry. Activities are shown as a percentage of the activity of PAK6-cat. For all kinase assays, SEM is shown, and values are calculated from more than three experiments. The 23-mer peptide significantly inhibits kinase activity. *P < 0.01, t test. (F) Src SH3 domain, but not β-PIX SH3 domain, increases PAK4-FL catalytic activity. PAK4-FL kinase activity toward MBP was assayed in the presence of either purified Src or β-PIX SH3 domain. MBP and SH3 domain loading are shown.
Fig. 6.
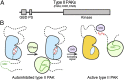
Regulation of type II PAKs. (A) Updated schematic diagram for type II PAK family members. The type II PAKs contain a GBD, an autoinhibitory pseudosubstrate (PS), and a C-terminal kinase domain. (B) Schematic showing regulation of type II PAKs. Activation loop-phosphorylated type II PAKs are autoinhibited by the pseudosubstrate sequence RPKP. GTPase binding to the GBD does not activate type II PAKs, but is important for subcellular localization. A second signal (e.g., an SH3 domain) releases pseudosubstrate autoinhibition to allow kinase activity, possibly through direct interaction with the pseudosubstrate sequence or another portion of the N terminus.
Similar articles
- Signaling, Regulation, and Specificity of the Type II p21-activated Kinases.
Ha BH, Morse EM, Turk BE, Boggon TJ. Ha BH, et al. J Biol Chem. 2015 May 22;290(21):12975-83. doi: 10.1074/jbc.R115.650416. Epub 2015 Apr 8. J Biol Chem. 2015. PMID: 25855792 Free PMC article. Review. - Substrate and inhibitor specificity of the type II p21-activated kinase, PAK6.
Gao J, Ha BH, Lou HJ, Morse EM, Zhang R, Calderwood DA, Turk BE, Boggon TJ. Gao J, et al. PLoS One. 2013 Oct 28;8(10):e77818. doi: 10.1371/journal.pone.0077818. eCollection 2013. PLoS One. 2013. PMID: 24204982 Free PMC article. - Rho family GTPase signaling through type II p21-activated kinases.
Chetty AK, Ha BH, Boggon TJ. Chetty AK, et al. Cell Mol Life Sci. 2022 Nov 19;79(12):598. doi: 10.1007/s00018-022-04618-2. Cell Mol Life Sci. 2022. PMID: 36401658 Free PMC article. Review. - Group I and II mammalian PAKs have different modes of activation by Cdc42.
Baskaran Y, Ng YW, Selamat W, Ling FT, Manser E. Baskaran Y, et al. EMBO Rep. 2012 Jun 29;13(7):653-9. doi: 10.1038/embor.2012.75. EMBO Rep. 2012. PMID: 22653441 Free PMC article. - Crystal structure of the SH3 domain of betaPIX in complex with a high affinity peptide from PAK2.
Hoelz A, Janz JM, Lawrie SD, Corwin B, Lee A, Sakmar TP. Hoelz A, et al. J Mol Biol. 2006 Apr 28;358(2):509-22. doi: 10.1016/j.jmb.2006.02.027. Epub 2006 Feb 28. J Mol Biol. 2006. PMID: 16527308
Cited by
- CDC42 binds PAK4 via an extended GTPase-effector interface.
Ha BH, Boggon TJ. Ha BH, et al. Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):531-536. doi: 10.1073/pnas.1717437115. Epub 2018 Jan 2. Proc Natl Acad Sci U S A. 2018. PMID: 29295922 Free PMC article. - The Cdc42 Effector Kinase PAK4 Localizes to Cell-Cell Junctions and Contributes to Establishing Cell Polarity.
Selamat W, Tay PL, Baskaran Y, Manser E. Selamat W, et al. PLoS One. 2015 Jun 11;10(6):e0129634. doi: 10.1371/journal.pone.0129634. eCollection 2015. PLoS One. 2015. PMID: 26068882 Free PMC article. - Signaling, Regulation, and Specificity of the Type II p21-activated Kinases.
Ha BH, Morse EM, Turk BE, Boggon TJ. Ha BH, et al. J Biol Chem. 2015 May 22;290(21):12975-83. doi: 10.1074/jbc.R115.650416. Epub 2015 Apr 8. J Biol Chem. 2015. PMID: 25855792 Free PMC article. Review. - p21-activated kinase signalling in pancreatic cancer: New insights into tumour biology and immune modulation.
Wang K, Baldwin GS, Nikfarjam M, He H. Wang K, et al. World J Gastroenterol. 2018 Sep 7;24(33):3709-3723. doi: 10.3748/wjg.v24.i33.3709. World J Gastroenterol. 2018. PMID: 30197477 Free PMC article. Review. - Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity.
Chen C, Ha BH, Thévenin AF, Lou HJ, Zhang R, Yip KY, Peterson JR, Gerstein M, Kim PM, Filippakopoulos P, Knapp S, Boggon TJ, Turk BE. Chen C, et al. Mol Cell. 2014 Jan 9;53(1):140-7. doi: 10.1016/j.molcel.2013.11.013. Epub 2013 Dec 26. Mol Cell. 2014. PMID: 24374310 Free PMC article.
References
- Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. - PubMed
- Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA121974/CA/NCI NIH HHS/United States
- GM102262/GM/NIGMS NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 GM102262/GM/NIGMS NIH HHS/United States
- GM079498/GM/NIGMS NIH HHS/United States
- P50 CA121974/CA/NCI NIH HHS/United States
- R01 GM079498/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous