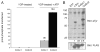Lack of Csk-mediated negative regulation in a unicellular SRC kinase - PubMed (original) (raw)
. 2012 Oct 16;51(41):8267-77.
doi: 10.1021/bi300965h. Epub 2012 Oct 1.
Affiliations
- PMID: 22998693
- PMCID: PMC3567254
- DOI: 10.1021/bi300965h
Lack of Csk-mediated negative regulation in a unicellular SRC kinase
Kira P Schultheiss et al. Biochemistry. 2012.
Abstract
Phosphotyrosine-based signaling plays a vital role in cellular communication in multicellular organisms. Unexpectedly, unicellular choanoflagellates (the closest phylogenetic group to metazoans) possess numbers of tyrosine kinases that are comparable to those in complex metazoans. Here, we have characterized tyrosine kinases from the filasterean Capsaspora owczarzaki, a unicellular protist representing the sister group to choanoflagellates and metazoans. Two Src-like tyrosine kinases have been identified in C. owczarzaki (CoSrc1 and CoSrc2), both of which have the arrangement of SH3, SH2, and catalytic domains seen in mammalian Src kinases. In Capsaspora cells, CoSrc1 and CoSrc2 localize to punctate structures in filopodia that may represent primordial focal adhesions. We have cloned, expressed, and purified both enzymes. CoSrc1 and CoSrc2 are active tyrosine kinases. Mammalian Src kinases are normally regulated in a reciprocal fashion by autophosphorylation in the activation loop (which increases activity) and by Csk-mediated phosphorylation of the C-terminal tail (which inhibits activity). Similar to mammalian Src kinases, the enzymatic activities of CoSrc1 and CoSrc2 are increased by autophosphorylation in the activation loop. We have identified a Csk-like kinase (CoCsk) in the genome of C. owczarzaki. We cloned, expressed, and purified CoCsk and found that it has no measurable tyrosine kinase activity. Furthermore, CoCsk does not phosphorylate or regulate CoSrc1 or CoSrc2 in cells or in vitro, and CoSrc1 and CoSrc2 are active in Capsaspora cell lysates. Thus, the function of Csk as a negative regulator of Src family kinases appears to have arisen with the emergence of metazoans.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1
Amino acid sequence alignment of C. owczarzaki CoSrc1 and CoSrc2, M. brevicollis MbSrc1, and human c-Src. The SH3 domain is boxed in red, the SH2 domain boxed in blue, and the kinase domain boxed in green. The orange circles represent conserved regulatory tyrosines: Tyr-416 in the activation loop and Tyr-527 in the C-terminal tail.
Figure 2
Phylogenetic tree of four cytoplasmic TK families. An alignment of 363 amino acid sites was used for the tree inference. The alignment covers SH3, SH2, and TK domains. GenBank accession numbers or Flybase IDs of sequences are given in brackets. Data for A. queenslandica, M. brevicollis, and C. owczarzaki were from the whole genome sequence. The numbers on branches indicate the statistical nodal support as obtained from 100 maximum likelihood bootstrap replicates using the WAG + Γ model of evolution.
Figure 3
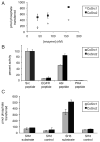
CoSrc1 and CoSrc2 are active tyrosine kinases. (A) Activity of CoSrc1 and CoSrc2 at varying enzyme concentrations. Enzyme activities were measured with 0.5 mM Src peptide substrate and 0.25 mM ATP using the phosphocellulose paper binding assay. Reactions proceeded for 5 min at 30 °C. (B) Substrate specificities of CoSrc1 and CoSrc2 were investigated using synthetic peptides incorporating recognition motifs from four protein kinases. Enzymes were incubated with 0.25 mM [_γ_-32P]ATP and 200 _μ_M peptide, and reactions proceeded for 20 min at 30 °C. Activities are presented relative to Src peptide (100%) for each enzyme. (C) Substrate targeting by CoSrc1 and CoSrc2. Enzymes (400 nM) were tested with synthetic peptide substrates containing SH3 or SH2 ligand sequences or matched controls (see Materials and Methods for peptide sequences). Reactions proceeded for 30 min at 30 °C, and mixtures were analyzed by the phosphocellulose paper assay. All assays were performed in duplicate, and error bars show standard deviations.
Figure 4
Autophosphorylation of CoSrc. (A) CoSrc1 and CoSrc2 were treated with immobilized GST-YOP for 30 min at 30 °C. The YOP was removed by centrifugation, and the mixture was assayed directly (left bars) or after incubation with ATP for 30 min at 30 °C (right bars). Activity measurements were performed in duplicate with the Src synthetic peptide and the phosphocellulose paper assay, as in Figure 2. (B) Whole cell lysates from untransfected SYF cells, or SYF cells expressing mammalian c-Src, CoSrc1, or CoSrc2, were analyzed by Western blotting with anti-phosphotyrosine antibody. The membrane was stripped and reprobed with anti-FLAG antibody to compare Src expression. The blots were quantitated by dividing the signal for autophosphorylated Src with the signal for total Src (ImageJ), and the value for c-Src was set to 1.0.
Figure 5
Amino acid sequence alignment of C. owczarzaki CoCsk, M. brevicollis MbCsk, and human Csk. The SH3 domain is boxed in red, the SH2 domain boxed in blue, and the kinase domain boxed in green. The orange circles represent sites that are important for interaction with Src.
Figure 6
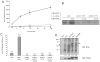
Activity of CoCsk is undetectable. (A) Purified CoCsk, M. brevicollis Csk (MbCsk), and human Csk (200 nM) were assayed with poly(Glu4Tyr) (1 mg/mL). Reaction mixtures contained 500 _μ_M [_γ_-32P]ATP. Activity was analyzed at various time points by scintillation counting. (B). CoCsk does not phosphorylate CoSrc in vitro. Purified CoCsk (400 nM) was incubated with purified CoSrc1 or CoSrc2 (activation loop mutants, 800 nM) in the presence of [_γ_-32P]ATP. Reactions were stopped by the addition of Laemmli buffer, and mixtures were analyzed by SDS-PAGE and autoradiography. (C) The bands shown in panel B were excised from the gel, dissolved, and analyzed by scintillation counting. (D) Whole cell lysates from SYF cells co-expressing the activation loop mutants (CoSrc1 Y447F or CoSrc2 Y471F) and CoCsk were analyzed by Western blotting with the anti-pTyr antibody. The filled arrow denotes the position of CoSrc1, and the empty arrow denotes the position of CoSrc2. The membrane was stripped and reprobed with anti-FLAG antibody to measure Src expression. The blots were quantitated by dividing the signal for autophosphorylated Src by the signal for total Src. Values in the lanes with CoCsk were compared to the values for CoSrc1 and CoSrc2 alone, which were each set to 1.0.
Figure 7
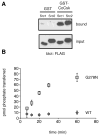
Biochemical studies of CoCsk. (A) Binding of CoSrc1 and CoSrc2 to CoCsk. GST-CoCsk (or GST control) was immobilized on glutathione-agarose beads. Lysates (0.5 mg) from SYF cells expressing CoSrc1 or CoSrc2 were incubated with the resins for 1 h at 4 °C. After being washed, bound proteins were analyzed by anti-FLAG Western blotting. For comparison, 5% of the lysates were analyzed by anti-FLAG Western blotting (bottom). (B) The G278N mutation partially restores CoCsk activity. Purified wild-type GST-CoCsk and GST-CoCsk-G278N were assayed with poly(Glu4Tyr) (1 mg/mL). Reaction mixtures contained 500 _μ_M [_γ_-32P]ATP. Activity was analyzed at various time points by scintillation counting.
Figure 8
CoCsk does not inhibit CoSrc Activity. (A) In vitro CoSrc kinase assays were performed in the presence or absence of purified CoCsk. Reaction mixtures contained 500 nM CoSrc1 or CoSrc2, CoCsk (600 nM), 500 _μ_M [_γ_-32P]ATP, and RCM-lysozyme (0.4 mg/mL) as a Src substrate. After 20 min at 30 °C, reaction mixtures were analyzed by scintillation counting, as described in Materials and Methods. For each enzyme, activity without CoCsk was normalized to 100%. (B) CoSrc1 or CoSrc2 was expressed in SYF cells in the presence and absence of CoCsk. CoSrc proteins were immunoprecipitated with immobilized FLAG antibody and incubated with Src substrate peptide and [_γ_-32P]ATP. After 20 min at 30 °C, activity was measured using the phosphocellulose paper assay. (C) Similar experiments were conducted to compare wild-type CoSrc1 and CoSrc2 (isolated from SYF cells co-expressing CoCsk) with mutant forms lacking C-terminal tyrosines. In each case, equivalent immunoprecipitation of CoSrc kinases was verified by anti-FLAG Western blotting (data not shown). (D) Western blot analyses of CoSrc1 (left) or CoSrc2 (right) activity (wild-type or C-terminal tail YF mutants) in the presence or absence of CoCsk. Whole cell lysates of SYF cells were analyzed by anti-pTyr blotting. The filled arrow denotes the position of CoSrc1; the empty arrow denotes the position of CoSrc2. Equal expression of CoSrc1 and CoSrc2 was confirmed by anti-FLAG blotting and equal expression of CoCsk by anti-V5 blotting. Quantitation was performed as described in the legend of Figure 6.
Figure 9
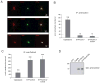
CoSrc localization and activity in C. owczarzaki cells. (A) Cells were fixed and immunostained with CoSrc1-, CoSrc2-, or actin-specific antibodies prior to fluorescence microscopy. Lectin stains the entire cell as well as the filopodia. (B) Anti-CoSrc1 immunoprecipitation reactions were conducted with lysates from Capsaspora cells or SYF cells overexpressing CoSrc1 in the presence or absence of CoCsk. Activity measurements were performed by the phosphocellulose paper assay with the Src synthetic peptide and [_γ_-32P]ATP. (C) Anti-CoSrc2 immunoprecipitation reactions were conducted with lysates from Capsaspora cells or SYF cells overexpressing CoSrc2 in the presence or absence of CoCsk. (D) Representative Western blot to compare the levels of CoSrc2 in Capsaspora and SYF cells.
Similar articles
- Regulation of Src and Csk nonreceptor tyrosine kinases in the filasterean Ministeria vibrans.
Schultheiss KP, Craddock BP, Suga H, Miller WT. Schultheiss KP, et al. Biochemistry. 2014 Mar 4;53(8):1320-9. doi: 10.1021/bi4016499. Epub 2014 Feb 18. Biochemistry. 2014. PMID: 24520931 Free PMC article. - Early emergence of negative regulation of the tyrosine kinase Src by the C-terminal Src kinase.
Taskinen B, Ferrada E, Fowler DM. Taskinen B, et al. J Biol Chem. 2017 Nov 10;292(45):18518-18529. doi: 10.1074/jbc.M117.811174. Epub 2017 Sep 22. J Biol Chem. 2017. PMID: 28939764 Free PMC article. - Novel mechanism of regulation of the non-receptor protein tyrosine kinase Csk: insights from NMR mapping studies and site-directed mutagenesis.
Shekhtman A, Ghose R, Wang D, Cole PA, Cowburn D. Shekhtman A, et al. J Mol Biol. 2001 Nov 16;314(1):129-38. doi: 10.1006/jmbi.2001.5126. J Mol Biol. 2001. PMID: 11724538 - Regulation of the SRC family kinases by Csk.
Okada M. Okada M. Int J Biol Sci. 2012;8(10):1385-97. doi: 10.7150/ijbs.5141. Epub 2012 Nov 1. Int J Biol Sci. 2012. PMID: 23139636 Free PMC article. Review. - Src protein-tyrosine kinase structure and regulation.
Roskoski R Jr. Roskoski R Jr. Biochem Biophys Res Commun. 2004 Nov 26;324(4):1155-64. doi: 10.1016/j.bbrc.2004.09.171. Biochem Biophys Res Commun. 2004. PMID: 15504335 Review.
Cited by
- Auto-thiophosphorylation activity of Src tyrosine kinase.
Cabail MZ, Chen EI, Koller A, Miller WT. Cabail MZ, et al. BMC Biochem. 2016 Jul 7;17(1):13. doi: 10.1186/s12858-016-0071-z. BMC Biochem. 2016. PMID: 27387461 Free PMC article. - Phosphotyrosine signalling and the origin of animal multicellularity.
Tong K, Wang Y, Su Z. Tong K, et al. Proc Biol Sci. 2017 Aug 16;284(1860):20170681. doi: 10.1098/rspb.2017.0681. Proc Biol Sci. 2017. PMID: 28768887 Free PMC article. - Src signaling in a low-complexity unicellular kinome.
Suga H, Miller WT. Suga H, et al. Sci Rep. 2018 Mar 29;8(1):5362. doi: 10.1038/s41598-018-23721-8. Sci Rep. 2018. PMID: 29599515 Free PMC article. - Temperature sensitivities of metazoan and pre-metazoan Src kinases.
Miller WT. Miller WT. Biochem Biophys Rep. 2020 Jun 10;23:100775. doi: 10.1016/j.bbrep.2020.100775. eCollection 2020 Sep. Biochem Biophys Rep. 2020. PMID: 32566764 Free PMC article. - The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition.
Ros-Rocher N, Pérez-Posada A, Leger MM, Ruiz-Trillo I. Ros-Rocher N, et al. Open Biol. 2021 Feb;11(2):200359. doi: 10.1098/rsob.200359. Epub 2021 Feb 24. Open Biol. 2021. PMID: 33622103 Free PMC article. Review.
References
- Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. - PubMed
- Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. - PubMed
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. - PubMed
- Kolibaba KS, Druker BJ. Protein tyrosine kinases and cancer. Biochim Biophys Acta. 1997;1333:F217–F248. - PubMed
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous