Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth - PubMed (original) (raw)
. 2012 Nov 8;491(7423):254-8.
doi: 10.1038/nature11465.
Kepeng Wang, Daniel Mucida, C Andrew Stewart, Bernd Schnabl, Dominik Jauch, Koji Taniguchi, Guann-Yi Yu, Christoph H Osterreicher, Kenneth E Hung, Christian Datz, Ying Feng, Eric R Fearon, Mohamed Oukka, Lino Tessarollo, Vincenzo Coppola, Felix Yarovinsky, Hilde Cheroutre, Lars Eckmann, Giorgio Trinchieri, Michael Karin
Affiliations
- PMID: 23034650
- PMCID: PMC3601659
- DOI: 10.1038/nature11465
Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth
Sergei I Grivennikov et al. Nature. 2012.
Abstract
Approximately 2% of colorectal cancer is linked to pre-existing inflammation known as colitis-associated cancer, but most develops in patients without underlying inflammatory bowel disease. Colorectal cancer often follows a genetic pathway whereby loss of the adenomatous polyposis coli (APC) tumour suppressor and activation of β-catenin are followed by mutations in K-Ras, PIK3CA and TP53, as the tumour emerges and progresses. Curiously, however, 'inflammatory signature' genes characteristic of colitis-associated cancer are also upregulated in colorectal cancer. Further, like most solid tumours, colorectal cancer exhibits immune/inflammatory infiltrates, referred to as 'tumour-elicited inflammation'. Although infiltrating CD4(+) T(H)1 cells and CD8(+) cytotoxic T cells constitute a positive prognostic sign in colorectal cancer, myeloid cells and T-helper interleukin (IL)-17-producing (T(H)17) cells promote tumorigenesis, and a 'T(H)17 expression signature' in stage I/II colorectal cancer is associated with a drastic decrease in disease-free survival. Despite its pathogenic importance, the mechanisms responsible for the appearance of tumour-elicited inflammation are poorly understood. Many epithelial cancers develop proximally to microbial communities, which are physically separated from immune cells by an epithelial barrier. We investigated mechanisms responsible for tumour-elicited inflammation in a mouse model of colorectal tumorigenesis, which, like human colorectal cancer, exhibits upregulation of IL-23 and IL-17. Here we show that IL-23 signalling promotes tumour growth and progression, and development of a tumoural IL-17 response. IL-23 is mainly produced by tumour-associated myeloid cells that are likely to be activated by microbial products, which penetrate the tumours but not adjacent tissue. Both early and late colorectal neoplasms exhibit defective expression of several barrier proteins. We propose that barrier deterioration induced by colorectal-cancer-initiating genetic lesions results in adenoma invasion by microbial products that trigger tumour-elicited inflammation, which in turn drives tumour growth.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures
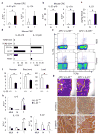
Figure 1. IL-23 controls CRC inflammation and tumorigenesis
a, b, RT–qPCR for IL-23p19 and IL-17A mRNAs from colorectal tumours (T) and matching normal (N) colons of (a) human CRC patients (n = 7, P = 0.037 for IL-23p19) or (b) CPC-APC mice (n = 8, P = 5 × 10−5, 3.6 × 10−4, respectively). c, IL-23 protein was measured by ELISA in supernatants of cultured tumours and normal tissues of CPC-APC mice (n = 5, P = 0.04). d, RT–qPCR analysis of sorted haematopoietic myeloid cells (CD45+TCRβ− CD11b+) from tumours of CPC-APC mice (n = 4; pooled); populations: CD11b+ = Gr1−F4/80low; Gr1+ = Gr1+F4/80− Gr1high = Gr1highF4/80− F4/80+ = Gr1−/dimF4/80+ and T cells (CD45+TCRβ+). e, Intracellular cytokine staining of phorbol myristate acetate and ionomycin re-stimulated cells (Live/Dead−CD45+ gate). f, Five-month-old _Il23_−/− and control CPC-APC mice were sacrificed and tumour numbers, size and load were determined (n = 7, P = 0.04, 0.03, 0.01, respectively). g, Tumour sections were stained with haematoxylin and eosin or phospho-STAT3 and β-catenin antibodies. h, Cytokine mRNA analysis by RT–qPCR in mesenteric lymph nodes (MLN), normal (N) and tumour (T) tissue of 5-month-old _Il23_−/− and control CPC-APC mice (n = 6, P = 0.044, 0.007, 0.045, respectively). Data represent averages ± s.e.m. *P < 0.05. Scale bars, 100μm.
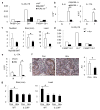
Figure 2. TLR-MyD88 signalling and commensal microflora promote cytokine expression and tumorigenesis
a–c, CPC-APC mice were transplanted with _Myd88_−/−, _Tlr2,4,9_−/− or control bone marrow (BM)and analysed 4 months later by RT–qPCR for cytokine mRNAs in sorted tumour myeloid cells (Live/Dead− TCRβ− CD11b+), representative of two independent experiments, each including four pooled mice (a) or in tumours and normal tissues (n = 5, P = 0.004, 0.032) (b). c, Tumour number, size and load in mice transplanted with indicated bone marrow (n = 5, P = 0.14, 0.048, 0.046, respectively). d–f, CPC-APC mice were treated with a cocktail of antibiotics (Abx) for 3 weeks. d, Myeloid cells were sorted from tumours and analysed by RT–qPCR, representative of two independent experiments, each including four pooled mice. e, IL-17A mRNA expression in normal and tumour tissues from the control and Abx-treated CPC-APC mice (n = 9, P = 0.003). f, Colon sections from mice were stained with phospho-STAT3 antibody, and intensity of staining was quantified (n = 8, P = 0.049). g, WT (Il23r+/−) or _Il23r_−/− CPC-APC mice were treated with antibiotics for 3.5 months and tumour size and load were determined (n = 5, P = 0.027 and 0.043). Data represent averages ± s.e.m. *P < 0.05. Scale bars, 50μm.
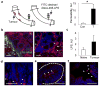
Figure 3. Colorectal tumours exhibit increased permeability to bacteria and their products
a, Colon segments of CPC-APC and control mice were clipped as indicated and their lumens injected with FITC-dextran or Alexa488-LPS. FITC fluorescence was measured in plasma 1 h later (n = 7, P = 0.017). b, Frozen colon sections prepared 30 min after Alexa488-LPS injection were stained with F4/80 antibody and DAPI and examined by fluorescent microscopy. c, Endotoxin was measured in portal blood of naive or tumour-bearing CPC-APC mice by Limulus bioassay (n = 9, P = 0.066). d–f, Colon sections from CPC-APC mice containing tumours (d) and early lesions (aberrant crypt foci) (e), and early human adenomas (f) were subjected to fluorescent in situ hybridization with eubacterial 16S-rRNA-specific Alexa594-labelled probe and stained with DAPI. Signals are indicated by the arrows. Data represent averages ± s.e.m. *P < 0.05. Scale bars, 50μm.
Figure 4. Colorectal tumours show defective mucin production and aberrant expression of junctional proteins
a, Colon sections from CPC-APC mice were stained with periodic acid-Schiff reagent, Oregon488-WGA or mucin 2 antibody. Normal (N) and tumour (T) areas are depicted. b, c, Sections of normal human colon and adjacent CRC specimens were stained with mucin-2-specific antibody (b) or JAM-A or JAM-B antibodies (c). d, Human adenomas were stained with WGA or JAM-A antibodies. Normal and tumour areas are marked. e, Sections of CPC-APC colons were stained with β-catenin antibody and analysed by light microscopy or WGA and JAM-A antibodies and analysed by confocal microscopy. f, Colon sections of tamoxifen-injected control or _Cdx2_ERT-Cre × _Apc_F/F mice were stained with JAM-A antibody or WGA. Normal (N) and APC-deleted areas are shown. Scale bars, 100μm.
References
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature Med. 2004;10:789–799. - PubMed
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. - PubMed
- Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
- Reichling T, et al. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer Res. 2005;65:166–176. - PubMed
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AA020703/AA/NIAAA NIH HHS/United States
- AI043477/AI/NIAID NIH HHS/United States
- R24 DK080506/DK/NIDDK NIH HHS/United States
- K99 DK088589/DK/NIDDK NIH HHS/United States
- K99-DK088589/DK/NIDDK NIH HHS/United States
- DK035108/DK/NIDDK NIH HHS/United States
- K08 DK081830/DK/NIDDK NIH HHS/United States
- R01 AI043477/AI/NIAID NIH HHS/United States
- R01 AI050265/AI/NIAID NIH HHS/United States
- DK080506/DK/NIDDK NIH HHS/United States
- P01 DK035108/DK/NIDDK NIH HHS/United States
- R37 AI043477/AI/NIAID NIH HHS/United States
- R01 CA082223/CA/NCI NIH HHS/United States
- R01CA082223/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous