Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21 - PubMed (original) (raw)
. 2012 Oct 23;109(43):17579-84.
doi: 10.1073/pnas.1211405109. Epub 2012 Oct 8.
Gillian Cowan, Adam J Mead, Sarah Filippi, Georg Bohn, Aristeidis Chaidos, Oliver Tunstall, Jerry K Y Chan, Mahesh Choolani, Phillip Bennett, Sailesh Kumar, Deborah Atkinson, Josephine Wyatt-Ashmead, Ming Hu, Michael P H Stumpf, Katerina Goudevenou, David O'Connor, Stella T Chou, Mitchell J Weiss, Anastasios Karadimitris, Sten Eirik Jacobsen, Paresh Vyas, Irene Roberts
Affiliations
- PMID: 23045701
- PMCID: PMC3491522
- DOI: 10.1073/pnas.1211405109
Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21
Anindita Roy et al. Proc Natl Acad Sci U S A. 2012.
Abstract
The 40-fold increase in childhood megakaryocyte-erythroid and B-cell leukemia in Down syndrome implicates trisomy 21 (T21) in perturbing fetal hematopoiesis. Here, we show that compared with primary disomic controls, primary T21 fetal liver (FL) hematopoietic stem cells (HSC) and megakaryocyte-erythroid progenitors are markedly increased, whereas granulocyte-macrophage progenitors are reduced. Commensurately, HSC and megakaryocyte-erythroid progenitors show higher clonogenicity, with increased megakaryocyte, megakaryocyte-erythroid, and replatable blast colonies. Biased megakaryocyte-erythroid-primed gene expression was detected as early as the HSC compartment. In lymphopoiesis, T21 FL lymphoid-primed multipotential progenitors and early lymphoid progenitor numbers are maintained, but there was a 10-fold reduction in committed PreproB-lymphoid progenitors and the functional B-cell potential of HSC and early lymphoid progenitor is severely impaired, in tandem with reduced early lymphoid gene expression. The same pattern was seen in all T21 FL samples and no samples had GATA1 mutations. Therefore, T21 itself causes multiple distinct defects in FL myelo- and lymphopoiesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
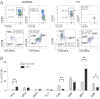
Fig. 1.
Perturbation of T21 FL HSC/progenitor frequency. (A) Representative plots from normal FL (n = 13) and T21 FL (n = 8) CD34+ cells gated on CD34+CD38+CD19− cells for CMP, MEP, and GMP, and on CD34+CD38lo/− cells for HSC, MPP, and LMPP. (B) Mean + SEM HSC and progenitor frequencies in normal (n = 13; light bars) and T21 (n = 8) FL (dark bars) showing increased HSC and MEP frequency and reduced CBP and GMP. **P < 0.01; *P < 0.05.
Fig. 2.
Increased clonogenicity and megakaryocyte/erythroid potential of normal and T21 FL HSC and progenitors. (A) Clonogenicity of flow-sorted HSC and progenitors from normal (n = 8) and T21 (n = 5) FL (mean + SEM). Cells (100 cells/mL) were plated in Methocult H4230 with IL-3, IL-6, IL-11, SCF, FLT3, GM-CSF, TPO, and EPO. Clonogenicity of T21 HSC, CMP, and MEP was increased compared to normal FL. **P < 0.01; *P < 0.05. (B) Lineage read out of clonogenic data in A showing increased MK and MK-erythroid (MkE) colonies from T21 FL HSC, MPP, CMP, and MEP, and Blast-My colonies from T21 FL LMPP compared to normal FL. T21 HSC generated increased absolute numbers of CFU-MK, MkE, BFU-E, Blast-E, and Blast-My compared to normal FL HSC and T21 CMP and MEP increased CFU-MK, MkE, and Blast-E (for quantitation see
Table S1
). (C) Representative colonies (Scale bars, 100 μm.) and (D) colony cytospins (Scale bars, 10 μm.) from normal (Left) and T21 (Right) FL clonogenic assays. (E) Only Blast-My and Blast-E had secondary replating activity. No tertiary replating was seen. There was no difference between replating activity of normal and T21 FL Blast-My or Blast-E.
Fig. 3.
Impaired lymphoid differentiation of T21 FL HSC, LMPP, and ELP. (A) Representative plots from normal (Left; n = 11) and T21 (Right; n = 6) FL showing reduced Pre-proB (CD34+CD19+CD10−) and ProB progenitors (CD34+CD19+CD10+). (B) Mean B-lymphoid progenitor frequencies in normal (light bars; n = 11) and T21 (n = 6) FL (dark bars) showing reduced PreProB and ProB progenitors in T21 FL. ***P < 0.001; *P < 0.02. (C) Representative plots showing reduced CD34−CD19+ cells in T21 compared to normal FL and summary data in normal (light bars, n = 5) and T21 FL (dark bars, n = 8). *P < 0.05. (D) Mean number of CD20+ cells/high-power fields from normal (n = 4; light bars) and T21 FL (n = 4; dark bars). (E) Differentiation of flow-sorted 100 HSC, LMPP, and ELP on MS5 stroma showing representative results on day 0 and day 14 (day 7 also shown for ELP) from normal (n = 3) and T21 FL (n = 4) and (Right) mean absolute number of CD19+ cells on day 14 from normal FL HSC, LMPP, and ELP (light bars) or T21 HSC, LMPP or ELP (dark bars).
Fig. 4.
(A–D) Altered gene expression in T21 FL HSC/progenitors Mean gene expression levels by quantitative RT-PCR from flow-sorted HSC/progenitors (50 cells in triplicate for each population) from normal (n = 5; light bars) and T21 FL (n = 3; dark bars) shown relative to GAPDH. Significant differences between T21 and normal FL are shown as *P < 0.05; **P < 0.01, and ***P < 0.001, using Bayesian analysis of differences in mean (see
SI Experimental Procedures
).
Comment in
- Leukaemia: Early changes.
McCarthy N. McCarthy N. Nat Rev Cancer. 2012 Dec;12(12):799. doi: 10.1038/nrc3403. Epub 2012 Nov 15. Nat Rev Cancer. 2012. PMID: 23151601 No abstract available.
Similar articles
- Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations.
Tunstall-Pedoe O, Roy A, Karadimitris A, de la Fuente J, Fisk NM, Bennett P, Norton A, Vyas P, Roberts I. Tunstall-Pedoe O, et al. Blood. 2008 Dec 1;112(12):4507-11. doi: 10.1182/blood-2008-04-152967. Epub 2008 Aug 8. Blood. 2008. PMID: 18689547 - Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs.
O'Byrne S, Elliott N, Rice S, Buck G, Fordham N, Garnett C, Godfrey L, Crump NT, Wright G, Inglott S, Hua P, Psaila B, Povinelli B, Knapp DJHF, Agraz-Doblas A, Bueno C, Varela I, Bennett P, Koohy H, Watt SM, Karadimitris A, Mead AJ, Ancliff P, Vyas P, Menendez P, Milne TA, Roberts I, Roy A. O'Byrne S, et al. Blood. 2019 Sep 26;134(13):1059-1071. doi: 10.1182/blood.2019001289. Epub 2019 Aug 5. Blood. 2019. PMID: 31383639 - SCL expression at critical points in human hematopoietic lineage commitment.
Zhang Y, Payne KJ, Zhu Y, Price MA, Parrish YK, Zielinska E, Barsky LW, Crooks GM. Zhang Y, et al. Stem Cells. 2005 Jun-Jul;23(6):852-60. doi: 10.1634/stemcells.2004-0260. Stem Cells. 2005. PMID: 15917481 - Delineating the cellular pathways of hematopoietic lineage commitment.
Luc S, Buza-Vidas N, Jacobsen SE. Luc S, et al. Semin Immunol. 2008 Aug;20(4):213-20. doi: 10.1016/j.smim.2008.07.005. Epub 2008 Aug 27. Semin Immunol. 2008. PMID: 18752972 Review. - Biological and molecular evidence for existence of lymphoid-primed multipotent progenitors.
Luc S, Buza-Vidas N, Jacobsen SE. Luc S, et al. Ann N Y Acad Sci. 2007 Jun;1106:89-94. doi: 10.1196/annals.1392.023. Epub 2007 Apr 18. Ann N Y Acad Sci. 2007. PMID: 17442777 Review.
Cited by
- Childhood leukaemia in Down's syndrome primed by blood-cell bias.
Malinge S. Malinge S. Nature. 2024 Oct;634(8032):40-42. doi: 10.1038/d41586-024-02785-9. Nature. 2024. PMID: 39322696 No abstract available. - Single-cell multi-omics map of human fetal blood in Down syndrome.
Marderstein AR, De Zuani M, Moeller R, Bezney J, Padhi EM, Wong S, Coorens THH, Xie Y, Xue H, Montgomery SB, Cvejic A. Marderstein AR, et al. Nature. 2024 Oct;634(8032):104-112. doi: 10.1038/s41586-024-07946-4. Epub 2024 Sep 25. Nature. 2024. PMID: 39322663 Free PMC article. - Down syndrome-associated leukaemias: current evidence and challenges.
Mason NR, Cahill H, Diamond Y, McCleary K, Kotecha RS, Marshall GM, Mateos MK. Mason NR, et al. Ther Adv Hematol. 2024 Jul 23;15:20406207241257901. doi: 10.1177/20406207241257901. eCollection 2024. Ther Adv Hematol. 2024. PMID: 39050114 Free PMC article. Review. - GATA1 in Normal and Pathologic Megakaryopoiesis and Platelet Development.
Takasaki K, Chou ST. Takasaki K, et al. Adv Exp Med Biol. 2024;1459:261-287. doi: 10.1007/978-3-031-62731-6_12. Adv Exp Med Biol. 2024. PMID: 39017848 Review. - Patterns of Aneuploidy and Signaling Consequences in Cancer.
Zhakula-Kostadinova N, Taylor AM. Zhakula-Kostadinova N, et al. Cancer Res. 2024 Aug 15;84(16):2575-2587. doi: 10.1158/0008-5472.CAN-24-0169. Cancer Res. 2024. PMID: 38924459 Free PMC article. Review.
References
- Parker SE, et al. National Birth Defects Prevention Network Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88(12):1008–1016. - PubMed
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nat Rev Genet. 2004;5(10):725–738. - PubMed
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355(9199):165–169. - PubMed
- Whitlock JA, et al. Clinical characteristics and outcome of children with Down syndrome and acute lymphoblastic leukemia: A Children's Cancer Group study. Blood. 2005;106(13):4043–4049. - PubMed
- Lange BJ, et al. Distinctive demography, biology, and outcome of acute myeloid leukemia and myelodysplastic syndrome in children with Down syndrome: Children’s Cancer Group Studies 2861 and 2891. Blood. 1998;91(2):608–615. - PubMed
Publication types
MeSH terms
Grants and funding
- G0501838/MRC_/Medical Research Council/United Kingdom
- G0801073/MRC_/Medical Research Council/United Kingdom
- G1002092/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/5/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials