The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells - PubMed (original) (raw)
. 2012 Oct 19;37(4):674-84.
doi: 10.1016/j.immuni.2012.09.008. Epub 2012 Oct 11.
Alan W Walker, Emilie Stolarczyk, James B Canavan, M Refik Gökmen, Ellen Marks, Ian Jackson, Ahmed Hashim, Michael A Curtis, Richard G Jenner, Jane K Howard, Julian Parkhill, Thomas T MacDonald, Graham M Lord
Affiliations
- PMID: 23063332
- PMCID: PMC3540260
- DOI: 10.1016/j.immuni.2012.09.008
The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells
Nick Powell et al. Immunity. 2012.
Abstract
Mice lacking the transcription factor T-bet in the innate immune system develop microbiota-dependent colitis. Here, we show that interleukin-17A (IL-17A)-producing IL-7Rα(+) innate lymphoid cells (ILCs) were potent promoters of disease in Tbx21(-/-)Rag2(-/-) ulcerative colitis (TRUC) mice. TNF-α produced by CD103(-)CD11b(+) dendritic cells synergized with IL-23 to drive IL-17A production by ILCs, demonstrating a previously unrecognized layer of cellular crosstalk between dendritic cells and ILCs. We have identified Helicobacter typhlonius as a key disease trigger driving excess TNF-α production and promoting colitis in TRUC mice. Crucially, T-bet also suppressed the expression of IL-7R, a key molecule involved in controlling intestinal ILC homeostasis. The importance of IL-7R signaling in TRUC disease was highlighted by the dramatic reduction in intestinal ILCs and attenuated colitis following IL-7R blockade. Taken together, these data demonstrate the mechanism by which T-bet regulates the complex interplay between mucosal dendritic cells, ILCs, and the intestinal microbiota.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1
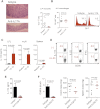
Chronic TRUC IBD Is Dependent on CD90+RORγt+CCR6+IL-7R+ ILCs (A) Intracellular cytokine expression by live, CD45+ cLP cells from TRUC mice following stimulation with PMA and ionomycin. (B) Phenotype of live, CD45+CD90hiIL-17A+ cells (red) in comparison with CD90−IL-17A− cells (black) from the mLN and cLP of TRUC mice following stimulation with PMA and ionomycin. (C) Intracellular cytokine expression by lineage− (CD11c−, NKp46−, Gr-1−) CD90hi ILCs from mLN of TRUC mice. Cells were stimulated with PMA and ionomycin. (D) IL-17A and CD90 expression in live, CD45+ cLP cells following in vivo administration of anti-CD90 or control antibody to TRUC mice (left panel). Cells were stimulated with PMA and ionomycin. Right panel shows absolute numbers of IL-17A-producing cells in the cLP of these TRUC mice following CD90 depletion (n = 4) or control mAb treatment (n = 5). Results show mean, and error bars represent SEM. Also see Figure S1A. (E) Colon micrographs and colitis scores following depleting anti-CD90 treatment in TRUC mice. Results show mean, and error bars represent SEM. Other clinical features are shown in Figure S1B. (F) Flow cytometry histogram showing the % CD90hi cells in the cLP of TRUC and TRUC × Il2rg −/− mice. Also see Figure S1C. (G) Colitis scores of TRUC (n = 14) and TRUC × Il2rg −/− (n = 5) mice. Results show mean, and error bars represent SEM. Other clinical features are shown in Figure S1D.
Figure 2
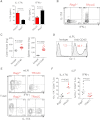
Chronic TRUC IBD Is Dependent on the IL-23:IL-17 Axis (A) Colon micrographs of TRUC mice following treatment with anti-IL17A (n = 7) or control antibody (n = 8). Also see Figure S2A for histology scores. (B) The proportion (%) of CD45+Gr-1hiCD11b+F4/80− granulocytes and F4/80+CD11b+ macrophages in the cLP of TRUC mice treated with anti-IL17A or isotype. A representative flow cytometry histogram is also shown (right panel). (C) IL-17A concentration in the supernatants of cultured cLPLs, mLN cells, and splenocytes from TRUC mice, in the presence (+IL-23) or absence (medium alone) of recombinant IL-23, measured by ELISA. Results show mean, and error bars represent SEM. (D) Intracellular cytokine expression in lin−CD90hi ILCs from mLN of TRUC mice following stimulation with IL-23. (E) IL-17A concentration in culture supernatants of explant organ culture of TRUC mice treated with anti-IL-23p19 or control antibody, measured by ELISA. Results show mean, and error bars represent SEM. (F) Colitis scores, colon mass, and spleen mass of TRUC mice treated with anti-IL-23p19 or control antibody. Results show mean, and error bars represent SEM.
Figure 3
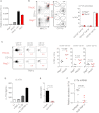
H. typhlonius Is a Key Component of the Intestinal Microbiota Driving IBD in TRUC Mice (A) Photomicrographs of the distal colon of 12-week-old TRUC mice with age matched, Tbx21 −/− Rag2 −/− mice that did not develop spontaneous colitis (TRnUC) (left panel). Macroscopic appearances of the colon of 16-week-old TRUC and TRnUC mice (right panel). (B) Colon and spleen mass in 12- to 16-week-old TRUC, TRnUC, and Rag2 −/− mice. ∗p < 0.0001 (TRUC versus TRnUC or Rag2 −/−). (C) Cluster dendrogram showing overall intestinal bacterial community membership clustering in TRUC and TRnUC mice. Families colored brown or yellow belong to the Bacteroidetes phylum; those in blue or purple belong to the Firmicutes phylum. SFB, segmented filamentous bacteria. Also see Figure S3B. (D) Agarose gel electrophoresis (2%) of PCR products following _H. typhlonius_-specific PCR performed on bacterial DNA isolated from fresh fecal samples from TRUC and TRnUC mice. The _H. typhlonius_-specific PCR product is 122 bp. (E) Colitis scores in TRnUC and Rag2 −/− mice following inoculation with pure cultures of H. typhlonius by oral gavage. Results show mean, and error bars represent SEM.
Figure 4
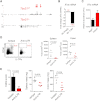
T-bet Is Required for Optimal Expression of IFN-γ by Intestinal ILCs, and in the Absence of T-bet ILCs Selectively Produced IL-17A (A) Cytokine concentrations in ex vivo explant cultures from Rag2 −/− (n = 5) and TRnUC (n = 5) mice following gavage with H. typhlonius, measured by ELISA. Results show mean, and error bars represent SEM. (B) Intracellular cytokine expression in live CD45+CD90hi cLP ILCs from Rag2 −/− and TRnUC mice in _H. typhlonius_-associated colitis. Cells were stimulated with PMA and ionomycin. (C) Organ mass in Rag2 −/− mice following treatment with agonistic CD40 mAbs or control antibody. Also see Figure S4A for other clinical features. (D) Flow cytometry histogram demonstrating the proportion of Gr-1hi granulocytes in the cLP of Rag2 −/− mice following anti-CD40 or control mAb administration. (E) Representative flow cytometry plot showing intracellular cytokine and T-bet expression in lin−CD90+ ILCs from mLN of Rag2 −/− and TRnUC mice following CD40 mAb treatment. (F) Proportions of IL-17A+ or IFN-γ+CD90+ ILCs in the cLP of Rag2 −/− or TRnUC mice following administration of anti-CD40.
Figure 5
CD103−CD11b+ Colonic DCs Are Major Producers of TNF-α, which Potentiates IL-23-Induced Innate IL-17 Expression (A) Real-time PCR measuring Tnfa transcripts in the colons of Rag2 −/−, TRnUC, TRnUC mice infected with H. typhlonius (+ HT), and TRUC mice. Results show mean, and error bars represent SEM. (B) DC subset frequency in the colon of TRnUC and Rag2 −/− mice following inoculation with H. typhlonius. Results show mean, and error bars represent SEM. (C) Spontaneous intracellular TNF-α production by different intestinal DC populations from Rag2 −/− and TRnUC mice following inoculation with H. typhlonius, showing representative flow cytometry plots (left panel) and statistical analysis (n = 5 per group) (right panel). (D) IL-17A production by unfractionated mLN cells from TRUC mice following incubation with the stimuli described. (-) indicates unstimulated cells. Results show mean, and error bars represent SEM. (E) Colitis scores in TRnUC mice inoculated with H. typhlonius following treatment with a blocking TNF-α mAb or control antibody. Results show mean, and error bars represent SEM. (F) Real-time PCR quantifying Il17a transcripts following treatment with a neutralizing TNF-α mAb or control isotype.
Figure 6
TRUC Disease Is Critically Dependent on IL-7R Signaling (A) UCSC Genome Browser image is shown for T-bet binding at the il7ra locus in Tbx21 +/+ versus Tbx21 −/− CD4+ Th1 cells following stimulation with PMA and ionomycin for 4 hr. Chromatin was immunoprecipitated, crosslinked, and prepared as described previously (Jenner et al., 2009). Samples were sequenced with an Illumina Genome Analyzer II-x. The vertical axis depicts the number of tags per million total sequences, with the genomic location running along the horizontal axis. The direction of transcription is indicated by arrows. Red arrows depict three sites of T-bet binding at the Il7ra locus. (B) Relative abundance of Il7ra mRNA in Tbx21 −/− Ifng −/− CD4+ T cells following retroviral transduction with empty vector (RV) or T-bet (T-bet RV). Cells were activated with PMA and ionomycin. (C) Relative abundance of Il7ra mRNA in fluorescence-activated cell-sorted Tbx21 −/− and Tbx21 +/+ CD90hi ILCs isolated from mLN of TRnUC and Rag2 −/− mice following induction of disease with anti-CD40. Results show mean, and error bars represent SEM. (D) Representative flow cytometry plot of splenic CD90+IL-7R+ ILCs (left panel) and absolute numbers (right panel) of ILCs in the spleen and colon of TRUC mice, following treatment with anti-IL7R or control antibody. (E) Colitis score (left panel), spleen mass (middle panel), and colon mass (right panel) in TRUC mice following treatment with anti-IL-7R or control antibody. Results show mean, and error bars represent SEM.
Similar articles
- Nod/Ripk2 signaling in dendritic cells activates IL-17A-secreting innate lymphoid cells and drives colitis in T-bet-/-.Rag2-/- (TRUC) mice.
Ermann J, Staton T, Glickman JN, de Waal Malefyt R, Glimcher LH. Ermann J, et al. Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):E2559-66. doi: 10.1073/pnas.1408540111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927559 Free PMC article. - T-Bet Controls Cellularity of Intestinal Group 3 Innate Lymphoid Cells.
Schroeder JH, Meissl K, Hromadová D, Lo JW, Neves JF, Howard JK, Helmby H, Powell N, Strobl B, Lord GM. Schroeder JH, et al. Front Immunol. 2021 Feb 2;11:623324. doi: 10.3389/fimmu.2020.623324. eCollection 2020. Front Immunol. 2021. PMID: 33603753 Free PMC article. - T-bet controls intestinal mucosa immune responses via repression of type 2 innate lymphoid cell function.
Garrido-Mesa N, Schroeder JH, Stolarczyk E, Gallagher AL, Lo JW, Bailey C, Campbell L, Sexl V, MacDonald TT, Howard JK, Grencis RK, Powell N, Lord GM. Garrido-Mesa N, et al. Mucosal Immunol. 2019 Jan;12(1):51-63. doi: 10.1038/s41385-018-0092-6. Epub 2018 Oct 24. Mucosal Immunol. 2019. PMID: 30356098 Free PMC article. - T-bet-/- RAG2-/- ulcerative colitis: the role of T-bet as a peacekeeper of host-commensal relationships.
Garrett WS, Glimcher LH. Garrett WS, et al. Cytokine. 2009 Oct-Nov;48(1-2):144-7. doi: 10.1016/j.cyto.2009.07.007. Epub 2009 Aug 8. Cytokine. 2009. PMID: 19666230 Free PMC article. Review. - Innate Lymphoid Cells in Intestinal Homeostasis and Inflammatory Bowel Disease.
Saez A, Gomez-Bris R, Herrero-Fernandez B, Mingorance C, Rius C, Gonzalez-Granado JM. Saez A, et al. Int J Mol Sci. 2021 Jul 16;22(14):7618. doi: 10.3390/ijms22147618. Int J Mol Sci. 2021. PMID: 34299236 Free PMC article. Review.
Cited by
- Innate Lymphocyte/Ly6C(hi) Monocyte Crosstalk Promotes Klebsiella Pneumoniae Clearance.
Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. Xiong H, et al. Cell. 2016 Apr 21;165(3):679-89. doi: 10.1016/j.cell.2016.03.017. Epub 2016 Mar 31. Cell. 2016. PMID: 27040495 Free PMC article. - Autoimmune host-microbiota interactions at barrier sites and beyond.
Ruff WE, Kriegel MA. Ruff WE, et al. Trends Mol Med. 2015 Apr;21(4):233-44. doi: 10.1016/j.molmed.2015.02.006. Epub 2015 Mar 11. Trends Mol Med. 2015. PMID: 25771098 Free PMC article. Review. - Microbiota-mediated inflammation and antimicrobial defense in the intestine.
Caballero S, Pamer EG. Caballero S, et al. Annu Rev Immunol. 2015;33:227-56. doi: 10.1146/annurev-immunol-032713-120238. Epub 2015 Jan 2. Annu Rev Immunol. 2015. PMID: 25581310 Free PMC article. Review. - Influence of orally fed a select mixture of Bacillus probiotics on intestinal T-cell migration in weaned MUC4 resistant pigs following Escherichia coli challenge.
Yang GY, Zhu YH, Zhang W, Zhou D, Zhai CC, Wang JF. Yang GY, et al. Vet Res. 2016 Jul 16;47(1):71. doi: 10.1186/s13567-016-0355-8. Vet Res. 2016. PMID: 27424033 Free PMC article. - Mixture of Five Fermented Herbs (Zhihuasi Tk) Alters the Intestinal Microbiota and Promotes the Growth Performance in Piglets.
Li Y, Sun T, Hong Y, Qiao T, Wang Y, Li W, Tang S, Yang X, Li J, Li X, Zhou Z, Xiao Y. Li Y, et al. Front Microbiol. 2021 Oct 26;12:725196. doi: 10.3389/fmicb.2021.725196. eCollection 2021. Front Microbiol. 2021. PMID: 34764942 Free PMC article.
References
- Araki A., Kanai T., Ishikura T., Makita S., Uraushihara K., Iiyama R., Totsuka T., Takeda K., Akira S., Watanabe M. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J. Gastroenterol. 2005;40:16–23. - PubMed
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., NIDDK IBD Genetics Consortium. Belgian-French IBD Consortium. Wellcome Trust Case Control Consortium Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT088747MA/WT_/Wellcome Trust/United Kingdom
- G0802068/MRC_/Medical Research Council/United Kingdom
- MR/J006742/1/MRC_/Medical Research Council/United Kingdom
- WT076964/WT_/Wellcome Trust/United Kingdom
- G0600081/MRC_/Medical Research Council/United Kingdom
- MR/J011118/1/MRC_/Medical Research Council/United Kingdom
- MR/K002996/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- DRF-2009-02-22/DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous