Estrogen receptor prevents p53-dependent apoptosis in breast cancer - PubMed (original) (raw)
Estrogen receptor prevents p53-dependent apoptosis in breast cancer
Shannon T Bailey et al. Proc Natl Acad Sci U S A. 2012.
Abstract
More than two-thirds of breast cancers express the estrogen receptor (ER) and depend on estrogen for growth and survival. Therapies targeting ER function, including aromatase inhibitors that block the production of estrogens and ER antagonists that alter ER transcriptional activity, play a central role in the treatment of ER+ breast cancers of all stages. In contrast to ER- breast cancers, which frequently harbor mutations in the p53 tumor suppressor, ER+ breast cancers are predominantly wild type for p53. Despite harboring wild-type p53, ER+ breast cancer cells are resistant to chemotherapy-induced apoptosis in the presence of estrogen. Using genome-wide approaches, we have addressed the mechanism by which ER antagonizes the proapoptotic function of p53. Interestingly, both ER agonists such as estradiol and the selective ER modulator (SERM) tamoxifen promote p53 antagonism. In contrast, the full ER antagonist fulvestrant blocks the ability of ER to inhibit p53-mediated cell death. This inhibition works through a mechanism involving the modulation of a subset of p53 and ER target genes that can predict the relapse-free survival of patients with ER+ breast cancer. These findings suggest an improved strategy for the treatment of ER+ breast cancer using antagonists that completely block ER action together with drugs that activate p53-mediated cell death.
Conflict of interest statement
Conflict of interest statement: M.B. serves as a consultant to Novartis and receives sponsored research support from Novartis and Pfizer.
Figures
Fig. 1.
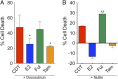
E2 and tamoxifen protect MCF7 cells against doxorubicin- and nutlin-mediated cell death. MCF7 cells were seeded in hormone-depleted medium containing vehicle (EtOH), E2, fulvestrant, or tamoxifen for 2 d and then treated with and without doxorubicin (A) or nutlin (B) for 3 d. Afterward, the cell viability was measured. The data shown were performed in triplicate and are representative of three independent experiments. These data demonstrate that E2 is protective against doxorubicin- and nutlin-mediated apoptosis in breast cancer cells. *P < 0.05 compared with doxorubicin alone treatment; **P < 0.005 compared with nutlin alone treatment.
Fig. 2.
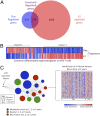
ER and p53 regulate a common subset of genes that is associated with poor outcome in patients with breast cancer. (A) We compared the 12-h doxorubicin-mediated gene expression profile in MCF7 cells, which was generated by using Affymetrix Human Genome U133A 2.0 arrays, with 12-h E2-mediated gene expression and found that 179 genes were shared by both stimuli. (B) We clustered these genes and found two main clusters: cluster 1 contained genes that were up-regulated by doxorubicin and down-regulated by E2, and cluster 2 comprised genes that were up-regulated by E2 and down-regulated by doxorubicin. (C) The 179 commonly regulated genes were interrogated by using Oncomine Concepts analysis (Compendia Biosciences) against publicly available primary breast tumor datasets, and significant associations were graphically represented in an interaction network by using Cytoscape (
). In this network, a node represents a dataset, and each edge represents a significant association with P < 0.01 and an odds ratio > 4. The 179 genes (shown in purple) demonstrated significant association with patient datasets exhibiting metastasis (blue circle), death (green circles), and recurrence (red circles) within 1, 3, or 5 y. The node size is proportional to the number of associated genes in each dataset. The van de Vijver dataset is shown on the right as an example of genes up-regulated in patients with recurrence at 3 y.
Fig. 3.
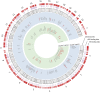
ER and p53 regulate a common set of gene targets. Circos plot demonstrating the ER and p53 cistromes, their relative genomic location, genes found within 100 kB of each transcription factor, and the clusters in which they reside. The outer hashes (shown in red) demonstrate the relative genomic location of the sites comprising the ER cistrome. The purple hashes display the location of the p53 cistrome. Genes possessing both an ER and p53 binding sites are also shown in red.
Fig. 4.
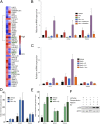
p53 activation combined with ER depletion leads to greater target gene expression. (A) Heat map demonstrating the relative expression level of each of the 36 regulated genes from gene expression microarray analysis of MCF7 cells treated with doxorubicin and E2. (B) ER loss leads to more efficient putative apoptotic gene expression. MCF7 cells were treated with doxorubicin alone or in combination with E2, fulvestrant, and tamoxifen to evaluate the expression of putative apoptosis target genes. Cotreatment with fulvestrant led to greater expression of these apoptotic genes, whereas treatment with tamoxifen inhibited in a manner similar to E2. (C) Similar effects were observed when treating with nutlin. (D and E) MCF7 cells were hormone starved and stimulated with doxorubicin (D) or nutlin (E). After a 4-h treatment, E2 was added, and a targeted p53 ChIP assay was performed. No difference was observed in the p53 recruitment level in response to doxorubicin (Left) or nutlin (Right) in the presence of E2. (F) Protein lysate from MCF7 cells treated under ChIP assay conditions were assayed by immunoblot for the level of p53 protein expression, and β-actin was blotted as a control.
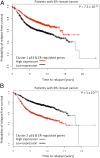
Fig. 5.
Kaplan-Meier analysis of patient datasets. Kaplan-Meier analysis reveals that the common regulated genes are predictors of relapse-free survival. To determine the role of these genes in predicting relapse-free survival in patients with ER+ breast cancer, we interrogated breast cancer datasets using the clusters 1 and 2 genes containing p53 and ER binding sites. (A) Kaplan-Meier analysis reveals that the cluster 1 genes that regulated by p53 and ER demonstrate better survival when they are highly expressed in patients with ER+ breast cancer. (B) The opposite result is observed when the p53 and ER-regulated cluster 2 genes are examined.
Similar articles
- Estrogen therapy induces an unfolded protein response to drive cell death in ER+ breast cancer.
Hosford SR, Shee K, Wells JD, Traphagen NA, Fields JL, Hampsch RA, Kettenbach AN, Demidenko E, Miller TW. Hosford SR, et al. Mol Oncol. 2019 Aug;13(8):1778-1794. doi: 10.1002/1878-0261.12528. Epub 2019 Jul 9. Mol Oncol. 2019. PMID: 31180176 Free PMC article. - Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer.
Sanchez CG, Ma CX, Crowder RJ, Guintoli T, Phommaly C, Gao F, Lin L, Ellis MJ. Sanchez CG, et al. Breast Cancer Res. 2011 Mar 1;13(2):R21. doi: 10.1186/bcr2833. Breast Cancer Res. 2011. PMID: 21362200 Free PMC article. - The proteasome inhibitor Bortezomib (Velcade) as potential inhibitor of estrogen receptor-positive breast cancer.
Thaler S, Thiede G, Hengstler JG, Schad A, Schmidt M, Sleeman JP. Thaler S, et al. Int J Cancer. 2015 Aug 1;137(3):686-97. doi: 10.1002/ijc.29404. Epub 2015 Jan 8. Int J Cancer. 2015. PMID: 25530422 - Selective estrogen modulators as an anticancer tool: mechanisms of efficiency and resistance.
Sengupta S, Jordan VC. Sengupta S, et al. Adv Exp Med Biol. 2008;630:206-19. doi: 10.1007/978-0-387-78818-0_13. Adv Exp Med Biol. 2008. PMID: 18637493 Review. - Changing role of the oestrogen receptor in the life and death of breast cancer cells.
Jordan VC, Osipo C, Schafer JM, Fox JE, Cheng D, Liu H. Jordan VC, et al. Breast. 2003 Dec;12(6):432-41. doi: 10.1016/s0960-9776(03)00149-8. Breast. 2003. PMID: 14659118 Review.
Cited by
- Elucidation of Mechanisms of Topotecan-Induced Cell Death in Human Breast MCF-7 Cancer Cells by Gene Expression Analysis.
Sinha BK, Tokar EJ, Bushel PR. Sinha BK, et al. Front Genet. 2020 Jul 17;11:775. doi: 10.3389/fgene.2020.00775. eCollection 2020. Front Genet. 2020. PMID: 32765594 Free PMC article. - Prognostic Value of the TP53 Mutation Location in Metastatic Breast Cancer as Detected by Next-Generation Sequencing.
Bai H, Yu J, Jia S, Liu X, Liang X, Li H. Bai H, et al. Cancer Manag Res. 2021 Apr 15;13:3303-3316. doi: 10.2147/CMAR.S298729. eCollection 2021. Cancer Manag Res. 2021. PMID: 33889023 Free PMC article. - MDM2 as a Rational Target for Intervention in CDK4/6 Inhibitor Resistant, Hormone Receptor Positive Breast Cancer.
Portman N, Chen J, Lim E. Portman N, et al. Front Oncol. 2021 Nov 3;11:777867. doi: 10.3389/fonc.2021.777867. eCollection 2021. Front Oncol. 2021. PMID: 34804982 Free PMC article. Review. - BCAS3 exhibits oncogenic properties by promoting CRL4A-mediated ubiquitination of p53 in breast cancer.
Zhou Z, Qiu R, Liu W, Yang T, Li G, Huang W, Teng X, Yang Y, Yu H, Yang Y, Wang Y. Zhou Z, et al. Cell Prolif. 2021 Aug;54(8):e13088. doi: 10.1111/cpr.13088. Epub 2021 Jul 9. Cell Prolif. 2021. PMID: 34240781 Free PMC article. - BRD4 Inhibition as a Strategy to Prolong the Response to Standard of Care in Estrogen Receptor-Positive Breast Cancer.
Elshazly AM, Sinanian MM, Neely V, Chakraborty E, Alshehri MA, McGrath MK, Harada H, Schoenlein PV, Gewirtz DA. Elshazly AM, et al. Cancers (Basel). 2023 Aug 11;15(16):4066. doi: 10.3390/cancers15164066. Cancers (Basel). 2023. PMID: 37627092 Free PMC article.
References
- Kanavos P. The rising burden of cancer in the developing world. Ann Oncol. 2006;17(Suppl 8):viii15–viii23. - PubMed
- Osborne CK, Zhao H, Fuqua SA. Selective estrogen receptor modulators: Structure, function, and clinical use. J Clin Oncol. 2000;18(17):3172–3186. - PubMed
- Osborne CK, Schiff R. Estrogen-receptor biology: Continuing progress and therapeutic implications. J Clin Oncol. 2005;23(8):1616–1622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA080111/CA/NCI NIH HHS/United States
- R01 DK074967/DK/NIDDK NIH HHS/United States
- R01 HG004069/HG/NHGRI NIH HHS/United States
- R01HG004069/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous