Resurrection of endogenous retroviruses in antibody-deficient mice - PubMed (original) (raw)
. 2012 Nov 29;491(7426):774-8.
doi: 10.1038/nature11599. Epub 2012 Oct 24.
Affiliations
- PMID: 23103862
- PMCID: PMC3511586
- DOI: 10.1038/nature11599
Resurrection of endogenous retroviruses in antibody-deficient mice
George R Young et al. Nature. 2012.
Abstract
The mammalian host has developed a long-standing symbiotic relationship with a considerable number of microbial species. These include the microbiota on environmental surfaces, such as the respiratory and gastrointestinal tracts, and also endogenous retroviruses (ERVs), comprising a substantial fraction of the mammalian genome. The long-term consequences for the host of interactions with these microbial species can range from mutualism to parasitism and are not always completely understood. The potential effect of one microbial symbiont on another is even less clear. Here we study the control of ERVs in the commonly used C57BL/6 (B6) mouse strain, which lacks endogenous murine leukaemia viruses (MLVs) able to replicate in murine cells. We demonstrate the spontaneous emergence of fully infectious ecotropic MLV in B6 mice with a range of distinct immune deficiencies affecting antibody production. These recombinant retroviruses establish infection of immunodeficient mouse colonies, and ultimately result in retrovirus-induced lymphomas. Notably, ERV activation in immunodeficient mice is prevented in husbandry conditions associated with reduced or absent intestinal microbiota. Our results shed light onto a previously unappreciated role for immunity in the control of ERVs and provide a potential mechanistic link between immune activation by microbial triggers and a range of pathologies associated with ERVs, including cancer.
Figures
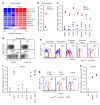
Figure 1. eMLV activation in antibody-deficient mice
a, Significantly upregulated (>4-fold) genes in CD11b+MHC-IIhiB220−Gr1− macrophages from _Rag1_−/− mice compared with macrophages from WT mice. Triplicate microarrays from cells isolated from 40 mice are shown. b, eMLV spliced env mRNA expression in the same cells as in a. Each symbol represents macrophages from 20 mice (_p_=0.024; paired Student’s t-test). c, eMLV spliced env mRNA expression in indicated organs from WT or _Rag1_−/− mice (spleen: _p_=0.020; small intestine: _p_=0.032; large intestine: _p_=0.004; lung: _p_= 0.001; muscle: _p_=0.016; and kidney: _p_=0.009; unpaired Student’s t-test). d, e, MLV SU expression in splenocytes (d) or in indicated cell types (e) from WT or _Rag1_−/− mice. f, eMLV spliced env mRNA expression in the spleens of the indicated strains (p<0.001 between WT and either _Ighm_−/− or _Ighm_−/− MD4 mice; one-way ANOVA). g, MLV SU expression in splenic lymphocytes from WT or _Ighm_−/− MD4 mice. h, eMLV spliced env mRNA expression in the spleens of the indicated strains (p<0.001 between WT and either _Myd88_−/− or _Tlr7_−/− mice; one-way ANOVA). In c, f and h, each symbol is an individual mouse. In d, e and g, plots are representative of 4 mice per group. In f and h, values above 103 were considered high and are indicated with red-filled symbols.
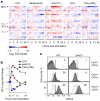
Figure 2. Retroviremia and leukaemias/lymphomas in antibody-deficient mice
a, Detection of infectious MLV (RARV-5/XG7) from the plasma of a representative _Rag1_−/− mouse by restoring infectivity of the green fluorescent protein (GFP)-expressing XG7 retroviral vector in the indicated cell type. Numbers within the plots denote the percentage of retrovirally-transduced (GFP+) cells. b, Fv1 tropism of RARVs isolated from 6- (R2-R4) or 25- (R5-R8) week-old healthy _Rag1_−/− mice, shown as the ratio of infectivity in B-3T3 to N-3T3 cells (B:N ratio). B- and N-tropic strains of Friend MLV (F-MLV) are shown for comparison. c, Amino acid residues of capsid positions 105-113 deduced from the nucleotide sequence of Emv2 and the same RARVs as in b. Dots indicate identities. d, Phylogenetic tree of the same RARVs as in b. The scale indicates the probability of base substitution per site. e, eMLV spliced env mRNA expression in the spleens of _Rag1_−/− mice or vertically-infected _Rag1_−/−_Emv2_−/− mice. Each dot is an individual mouse (p<0.001; unpaired Student’s t-test). Values above 103 were considered high and are indicated with red-filled symbols. f, MLV SU expression in splenocytes from _Rag1_−/− or vertically-infected _Rag1_−/−_Emv2_−/− mice (representative of 9 mice per group). g, eMLV DNA copy numbers per haploid genome, determined by qPCR for the pol or ecotropic env gene, in DNA from the spleens of healthy _Rag1_−/− (right) or vertically-infected _Rag1_−/− _Emv2_−/− mice (left). Symbols represent individual mice, grouped according to their age. The sensitivity limit of this PCR method was determined as a median of 0.0003 copies per haploid genome, using _Emv2_−/− mice. eMLV DNA copy numbers for _Rag1_−/− mice include Emv2 (1/N). h, Tumour (leukaemias/lymphomas) incidence in cohorts of WT (n=37), _Rag1_−/− (n=38) or vertically-infected _Rag1_−/−_Emv2_−/− mice (n=23) at the NIMR SPF facility (p<0.000001 between WT and _Rag1_−/− mice; _p_=0.00025 between WT and _Rag1_−/−_Emv2_−/− mice; Log-Rank Survival analysis).
Figure 3. Murine ERV activation by microbial products
a, ERV/RE-reporting probeset (Supplementary Table 3) signals in a publicly-available Affymetrix HT Mouse Genome 430A microarray dataset (E-GEOD-17721) of WT B6 bone marrow-derived dendritic cells following stimulation with microbial products. Black arrows indicate the probesets that are significantly regulated (p<0.05) more than 2-fold by at least one stimulus. _Mela (Emv2)_-specific probesets are also indicated by grey arrows for comparison. b, Mean log2 fold-change in MLV-reporting probeset in the same dataset. c, MLV SU expression in WT or _Emv2_−/− splenocytes before (open histograms) and after stimulation with 10 μg/ml LPS for 48 hrs (grey-shaded histograms), according to Forward scatter (Fsc) and CD19 expression. Numbers with the plots denote the percentage of cells within each gate and represent 2 donors each analysed in duplicate.
Figure 4. eMLV activation in antibody-deficiency depends on husbandry conditions
a, eMLV spliced env mRNA expression in the spleens of _Rag1_−/− mice on neutral pH (SPF) or acidified water (pH 2.5) at NIMR, on acidified water (pH 2.8) at JAX, on neutral pH at RCHCI or in germ-free (GF) facilities at UMICH (p<0.016 between _Rag1_−/− mice at NIMR SPF and all other groups; one-way ANOVA). b, eMLV spliced env mRNA expression in the spleens of _Ighm_−/−, _Myd88_−/− or _Tlr7_−/− mice on neutral pH water (SPF) at NIMR or on acidified water (pH 2.8) at JAX (_p_=0.005 and _p_=0.029 for _Ighm_−/− and _Tlr7_−/− mice, respectively; unpaired Student’s t-test). Each dot is an individual mouse and values above 103 were considered high and are indicated with red-filled symbols.
Comment in
- Antiviral immunity: Immune control of endogenous retroviruses.
Leavy O. Leavy O. Nat Rev Immunol. 2012 Dec;12(12):805. doi: 10.1038/nri3355. Nat Rev Immunol. 2012. PMID: 23175217 No abstract available.
Similar articles
- Xenotropic Mouse Gammaretroviruses Isolated from Pre-Leukemic Tissues Include a Recombinant.
Bamunusinghe D, Skorski M, Buckler-White A, Kozak CA. Bamunusinghe D, et al. Viruses. 2018 Aug 9;10(8):418. doi: 10.3390/v10080418. Viruses. 2018. PMID: 30096897 Free PMC article. - Distribution of endogenous gammaretroviruses and variants of the Fv1 restriction gene in individual mouse strains and strain subgroups.
Skorski M, Bamunusinghe D, Liu Q, Shaffer E, Kozak CA. Skorski M, et al. PLoS One. 2019 Jul 10;14(7):e0219576. doi: 10.1371/journal.pone.0219576. eCollection 2019. PLoS One. 2019. PMID: 31291374 Free PMC article. - Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor.
Bamunusinghe D, Liu Q, Lu X, Oler A, Kozak CA. Bamunusinghe D, et al. J Virol. 2013 Sep;87(17):9845-55. doi: 10.1128/JVI.01264-13. Epub 2013 Jul 3. J Virol. 2013. PMID: 23824809 Free PMC article. - Origins of the endogenous and infectious laboratory mouse gammaretroviruses.
Kozak CA. Kozak CA. Viruses. 2014 Dec 26;7(1):1-26. doi: 10.3390/v7010001. Viruses. 2014. PMID: 25549291 Free PMC article. Review. - Role of endogenous retroviruses in autoimmune diseases.
Perl A. Perl A. Rheum Dis Clin North Am. 2003 Feb;29(1):123-43, vii. doi: 10.1016/s0889-857x(02)00098-4. Rheum Dis Clin North Am. 2003. PMID: 12635504 Review.
Cited by
- Repetitive Sequence Stability in Embryonic Stem Cells.
Shi G, Pang Q, Lin Z, Zhang X, Huang K. Shi G, et al. Int J Mol Sci. 2024 Aug 13;25(16):8819. doi: 10.3390/ijms25168819. Int J Mol Sci. 2024. PMID: 39201503 Free PMC article. Review. - Adaptive immunity to retroelements promotes barrier integrity.
Wells AC, Lima-Junior DS, Link VM, Smelkinson M, Krishnamurthy SR, Chi L, Segrist E, Rivera CA, Teijeiro A, Bouladoux N, Belkaid Y. Wells AC, et al. bioRxiv [Preprint]. 2024 Aug 9:2024.08.09.606346. doi: 10.1101/2024.08.09.606346. bioRxiv. 2024. PMID: 39149266 Free PMC article. Preprint. - Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element.
Williams ZH, Imedio AD, Gaucherand L, Lee DC, Mostafa SM, Phelan JP, Coffin JM, Johnson WE. Williams ZH, et al. Elife. 2024 Jul 22;13:e80216. doi: 10.7554/eLife.80216. Elife. 2024. PMID: 39037763 Free PMC article. - Comprehensive Identification and Characterization of HML-9 Group in Chimpanzee Genome.
Chen M, Yang C, Zhai X, Wang C, Liu M, Zhang B, Guo X, Wang Y, Li H, Liu Y, Han J, Wang X, Li J, Jia L, Li L. Chen M, et al. Viruses. 2024 May 31;16(6):892. doi: 10.3390/v16060892. Viruses. 2024. PMID: 38932184 Free PMC article. - T-bet+ B cells are activated by and control endogenous retroviruses through TLR-dependent mechanisms.
Rauch E, Amendt T, Lopez Krol A, Lang FB, Linse V, Hohmann M, Keim AC, Kreutzer S, Kawengian K, Buchholz M, Duschner P, Grauer S, Schnierle B, Ruhl A, Burtscher I, Dehnert S, Kuria C, Kupke A, Paul S, Liehr T, Lechner M, Schnare M, Kaufmann A, Huber M, Winkler TH, Bauer S, Yu P. Rauch E, et al. Nat Commun. 2024 Feb 9;15(1):1229. doi: 10.1038/s41467-024-45201-6. Nat Commun. 2024. PMID: 38336876 Free PMC article.
References
- Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. - PubMed
- King SR, Berson BJ, Risser R. Mechanism of interaction between endogenous ecotropic murine leukemia viruses in (BALB/c X C57BL/6) hybrid cells. Virology. 1988;162:1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U.1175.02.006.00007(81330)/MRC_/Medical Research Council/United Kingdom
- MC_U117512710/MRC_/Medical Research Council/United Kingdom
- U117512710/MRC_/Medical Research Council/United Kingdom
- U117581330/MRC_/Medical Research Council/United Kingdom
- U.1175.02.005.00005(60891)/MRC_/Medical Research Council/United Kingdom
- MC_U117581330/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases