Haploinsufficiency at the human IFNGR2 locus contributes to mycobacterial disease - PubMed (original) (raw)
. 2013 Feb 15;22(4):769-81.
doi: 10.1093/hmg/dds484. Epub 2012 Nov 16.
Guillaume Vogt, Yuval Itan, Anna Macura-Biegun, Anna Szaflarska, Danuta Kowalczyk, Ariane Chapgier, Avinash Abhyankar, Dieter Furthner, Claudia Djambas Khayat, Satoshi Okada, Vanessa L Bryant, Dusan Bogunovic, Alexandra Kreins, Marcela Moncada-Vélez, Mélanie Migaud, Sulaiman Al-Ajaji, Saleh Al-Muhsen, Steven M Holland, Laurent Abel, Capucine Picard, Damien Chaussabel, Jacinta Bustamante, Jean-Laurent Casanova, Stéphanie Boisson-Dupuis
Affiliations
- PMID: 23161749
- PMCID: PMC3554203
- DOI: 10.1093/hmg/dds484
Haploinsufficiency at the human IFNGR2 locus contributes to mycobacterial disease
Xiao-Fei Kong et al. Hum Mol Genet. 2013.
Abstract
Mendelian susceptibility to mycobacterial diseases (MSMD) is a rare syndrome, the known genetic etiologies of which impair the production of, or the response to interferon-gamma (IFN-γ). We report here a patient (P1) with MSMD whose cells display mildly impaired responses to IFN-γ, at levels, however, similar to those from MSMD patients with autosomal recessive (AR) partial IFN-γR2 or STAT1 deficiency. Whole-exome sequencing (WES) and Sanger sequencing revealed only one candidate variation for both MSMD-causing and IFN-γ-related genes. P1 carried a heterozygous frame-shift IFNGR2 mutation inherited from her father. We show that the mutant allele is intrinsically loss-of-function and not dominant-negative, suggesting haploinsufficiency at the IFNGR2 locus. We also show that Epstein-Barr virus transformed B lymphocyte cells from 10 heterozygous relatives of patients with AR complete IFN-γR2 deficiency respond poorly to IFN-γ, in some cases as poorly as the cells of P1. Naive CD4(+) T cells and memory IL-4-producing T cells from these individuals also responded poorly to IFN-γ, whereas monocytes and monocyte-derived macrophages (MDMs) did not. This is consistent with the lower levels of expression of IFN-γR2 in lymphoid than in myeloid cells. Overall, MSMD in this patient is probably due to autosomal dominant (AD) IFN-γR2 deficiency, resulting from haploinsufficiency, at least in lymphoid cells. The clinical penetrance of AD IFN-γR2 deficiency is incomplete, possibly due, at least partly, to the variability of cellular responses to IFN-γ in these individuals.
Figures
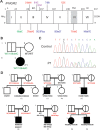
Figure 1.
Pedigree and mutations of IFNGR2. (A) Schematic presentation of the mutations identified in the IFNGR2 gene. The leader sequence (L, 1–22), extracellular domain (EC, 23–248), transmembrane domain (TM, 249–272) and intracellular domain (IC, 273–337) are indicated. Mutations marked in red cause complete AR IFN-γR2 deficiency with no detectable expression of IFN-γR2 at the cell surface. The mutations marked in blue cause complete AR IFN-γR2 deficiency with detectable surface expression of a non-functional IFN-γR2. With the antibody now available, the cells of patients carrying the 382–387dup and 663del27 mutations (26) were shown to have impaired, but detectable IFN-γR2 expression at the surface of Epstein-Barr virus transformed B lymphocyte (EBV-B) cells. The mutation marked in purple cause partial AR IFN-γR2 deficiency. The mutation marked in green is the heterozygous mutation reported here. (B) Family pedigree of P1. (C) Electropherogram showing the heterozygous IFNGR2 186delC mutation found in P1. (D) Pedigree of the families with IFN-γR2 deficiency recruited for this study. The IFNGR2 genotype of the heterozygous individuals included is shown in red.
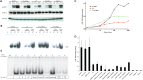
Figure 2.
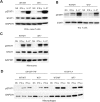
P1's IFN-γ response and exploration of a possible dominant-negative effect. EBV-B cells from a healthy control, P1, P1's father and mother were stimulated with IFN-γ or IFN-α and subjected to western blot analysis for STAT1, Tyr701 phosphorylated STAT1 (A) or to EMSA for the assessment of GAF DNA-binding activities (B). (C) The GAF DNA-binding activities of EBV-B cells stimulated with various concentrations of IFN-γ were compared between a healthy control, P1 and P1's father. (D) GAF DNA-binding activities in the EBV-B cells were compared between a healthy control, individuals heterozygous for IFNGR1 or STAT1, P1 and patients with various IFNGR1, IFNGR2 and STAT1 mutations resulting in partial or complete deficiency, after stimulation with IFN-γ. The genotype of each cell line is written for the corresponding gene. (E) SV40-transformed fibroblasts from a healthy control were transfected with various plasmids encoding the WT or a mutant form of IFN-γR2. The various mutations included 186delC, 663del27 and 791delG, 382–87dup. Cells were also transfected with an IFNGR1 818del4 or STAT1 L706S expression plasmid as a positive control, for the detection of dominant-negative effects on IFN-γ signaling.
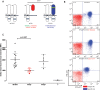
Figure 3.
IFN-γR2 expression in heterozygous IFNGR2 cells. (A) Schematic diagram of IFN-γR2 structures. In WT/WT healthy controls, the protein is encoded by two WT alleles. WT/E− individuals have a heterozygous loss-of-function mutation preventing the expression of the receptor on the cell surface. WT/E+ individuals have a loss-of-function mutation changing the structure of IFN-γR2, but with protein detectable on the cell surface. (B) Monocytes from a healthy control, an individual carrying a heterozygous IFNGR2 218delAA mutation and an individual carrying a heterozygous IFNGR1 202–1G>T mutation were incubated with an antibody recognizing the extracellular domain of IFN-γR1 or IFN-γR2. The red plot corresponds to the isotype control. The blue plot corresponds to the specific antibody for IFN-γR1 and IFN-γR2. (C) Delta mean fluorescence intensity (ΔMFI) of IFN-γR2 in EBV-B cells was compared between healthy controls, WT/E−, WT/E+ and 218delAA/218delAA patients. The solid red circles indicate the ΔMFI of the parent with a heterozygous 791delG mutation. Student's _t_-test was used to assess the significance of differences between two groups. Each experiment was carried out at least three times.
Figure 4.
Haploinsufficiency in EBV-B cells from heterozygous IFNGR2 individuals. (A) Tyr701 STAT1 phosphorylation was quantified by FACS in EBV-B cells after stimulation with 105 IU/ml IFN-γ or IFN-α for 30 min. These experiments were carried out twice. (B) GAF DNA-binding activities in EBV-B cells were compared between healthy controls, heterozygous IFNGR1 individuals and heterozygous IFNGR2 individuals following stimulation with various concentrations of IFN-γ or IFN-α, as indicated. The dot indicates the relative value of GAF DNA-binding activity for a given cell with respect to the same healthy control. The solid red circles and squares indicate the values for the parent with a heterozygous 791delG mutation. The solid green circles and squares indicate the values for P1. (C) P1's GAF DNA-binding activity was compared with that of the other individuals heterozygous for IFNGR2.
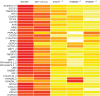
Figure 5.
Microarray analysis of ISGs in EBV-B cells. EBV-B cells from three healthy controls, P1 and patients with complete STAT1, IFN-γR1 and IFN-γR2 deficiencies were stimulated by incubation with 104 IU/ml IFN-γ for 2 h. Red colors represent high stimulated/basal expression fold induction level. Whole-transcriptome analysis was then carried out on mRNA extracted from EBV-B cells.
Figure 6.
Haploinsufficiency in the primary immune cells from heterozygous IFNGR2 individuals. (A) Naive CD4+ T cells, (B) IL-4-producing T helper cells, (C) monocytes (D) MDMs generated in the presence of either M-CSF + IL-4 or GM-CSF + TNF-α were stimulated by incubation with IFN-γ, IFN-α or IL-27 for 15 min. Western blots were carried out to evaluate STAT1 phosphorylation. These experiments were carried out at least twice.
Similar articles
- A purely quantitative form of partial recessive IFN-γR2 deficiency caused by mutations of the initiation or second codon.
Oleaga-Quintas C, Deswarte C, Moncada-Vélez M, Metin A, Krishna Rao I, Kanık-Yüksek S, Nieto-Patlán A, Guérin A, Gülhan B, Murthy S, Özkaya-Parlakay A, Abel L, Martínez-Barricarte R, Pérez de Diego R, Boisson-Dupuis S, Kong XF, Casanova JL, Bustamante J. Oleaga-Quintas C, et al. Hum Mol Genet. 2018 Nov 15;27(22):3919-3935. doi: 10.1093/hmg/ddy275. Hum Mol Genet. 2018. PMID: 31222290 Free PMC article. - Enhanced osteoclastogenesis in patients with MSMD due to impaired response to IFN-γ.
Tsumura M, Miki M, Mizoguchi Y, Hirata O, Nishimura S, Tamaura M, Kagawa R, Hayakawa S, Kobayashi M, Okada S. Tsumura M, et al. J Allergy Clin Immunol. 2022 Jan;149(1):252-261.e6. doi: 10.1016/j.jaci.2021.05.018. Epub 2021 Jun 24. J Allergy Clin Immunol. 2022. PMID: 34176646 - Patient iPSC-Derived Macrophages to Study Inborn Errors of the IFN-γ Responsive Pathway.
Haake K, Neehus AL, Buchegger T, Kühnel MP, Blank P, Philipp F, Oleaga-Quintas C, Schulz A, Grimley M, Goethe R, Jonigk D, Kalinke U, Boisson-Dupuis S, Casanova JL, Bustamante J, Lachmann N. Haake K, et al. Cells. 2020 Feb 19;9(2):483. doi: 10.3390/cells9020483. Cells. 2020. PMID: 32093117 Free PMC article. - Mendelian susceptibility to mycobacterial diseases: state of the art.
Noma K, Mizoguchi Y, Tsumura M, Okada S. Noma K, et al. Clin Microbiol Infect. 2022 Nov;28(11):1429-1434. doi: 10.1016/j.cmi.2022.03.004. Epub 2022 Mar 11. Clin Microbiol Infect. 2022. PMID: 35283318 Review. - The genetic heterogeneity of mendelian susceptibility to mycobacterial diseases.
Al-Muhsen S, Casanova JL. Al-Muhsen S, et al. J Allergy Clin Immunol. 2008 Dec;122(6):1043-51; quiz 1052-3. doi: 10.1016/j.jaci.2008.10.037. J Allergy Clin Immunol. 2008. PMID: 19084105 Review.
Cited by
- A novel homozygous p.R1105X mutation of the AP4E1 gene in twins with hereditary spastic paraplegia and mycobacterial disease.
Kong XF, Bousfiha A, Rouissi A, Itan Y, Abhyankar A, Bryant V, Okada S, Ailal F, Bustamante J, Casanova JL, Hirst J, Boisson-Dupuis S. Kong XF, et al. PLoS One. 2013;8(3):e58286. doi: 10.1371/journal.pone.0058286. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472171 Free PMC article. - A purely quantitative form of partial recessive IFN-γR2 deficiency caused by mutations of the initiation or second codon.
Oleaga-Quintas C, Deswarte C, Moncada-Vélez M, Metin A, Krishna Rao I, Kanık-Yüksek S, Nieto-Patlán A, Guérin A, Gülhan B, Murthy S, Özkaya-Parlakay A, Abel L, Martínez-Barricarte R, Pérez de Diego R, Boisson-Dupuis S, Kong XF, Casanova JL, Bustamante J. Oleaga-Quintas C, et al. Hum Mol Genet. 2018 Nov 15;27(22):3919-3935. doi: 10.1093/hmg/ddy275. Hum Mol Genet. 2018. PMID: 31222290 Free PMC article. - Severe viral respiratory infections in children with IFIH1 loss-of-function mutations.
Asgari S, Schlapbach LJ, Anchisi S, Hammer C, Bartha I, Junier T, Mottet-Osman G, Posfay-Barbe KM, Longchamp D, Stocker M, Cordey S, Kaiser L, Riedel T, Kenna T, Long D, Schibler A, Telenti A, Tapparel C, McLaren PJ, Garcin D, Fellay J. Asgari S, et al. Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8342-8347. doi: 10.1073/pnas.1704259114. Epub 2017 Jul 17. Proc Natl Acad Sci U S A. 2017. PMID: 28716935 Free PMC article. - Phenotypic and immune functional profiling of patients with suspected Mendelian Susceptibility to Mycobacterial Disease in South Africa.
van Coller A, Glanzmann B, Cornelissen H, Möller M, Kinnear C, Esser M, Glashoff R. van Coller A, et al. BMC Immunol. 2021 Sep 13;22(1):62. doi: 10.1186/s12865-021-00452-6. BMC Immunol. 2021. PMID: 34517836 Free PMC article. - A post-GWAS analysis of predicted regulatory variants and tuberculosis susceptibility.
Uren C, Henn BM, Franke A, Wittig M, van Helden PD, Hoal EG, Möller M. Uren C, et al. PLoS One. 2017 Apr 6;12(4):e0174738. doi: 10.1371/journal.pone.0174738. eCollection 2017. PLoS One. 2017. PMID: 28384278 Free PMC article.
References
- Casanova J.L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 2002;20:581–620. - PubMed
- Casanova J.L., Abel L. The human model: a genetic dissection of immunity to infection in natural conditions. Nat. Rev. Immunol. 2004;4:55–66. - PubMed
- Fortin A., Abel L., Casanova J.L., Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu. Rev. Genomics Hum. Genet. 2007;8:163–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000043/TR/NCATS NIH HHS/United States
- R37 AI095983/AI/NIAID NIH HHS/United States
- 8UL1TR000043/TR/NCATS NIH HHS/United States
- R01 AI089970/AI/NIAID NIH HHS/United States
- 5R01AI089970-02/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous