PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity - PubMed (original) (raw)
PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity
Khalil Choucair et al. BMC Cancer. 2012.
Abstract
Background: Prostate cancer (PCa), a leading cause of cancer death in North American men, displays a broad range of clinical outcome from relatively indolent to lethal metastatic disease. Several genomic alterations have been identified in PCa which may serve as predictors of progression. PTEN, (10q23.3), is a negative regulator of the phosphatidylinositol 3-kinase (PIK3)/AKT survival pathway and a tumor suppressor frequently deleted in PCa. The androgen receptor (AR) signalling pathway is known to play an important role in PCa and its blockade constitutes a commonly used treatment modality. In this study, we assessed the deletion status of PTEN along with AR expression levels in 43 primary PCa specimens with clinical follow-up.
Methods: Fluorescence In Situ Hybridization (FISH) was done on formalin fixed paraffin embedded (FFPE) PCa samples to examine the deletion status of PTEN. AR expression levels were determined using immunohistochemistry (IHC).
Results: Using FISH, we found 18 cases of PTEN deletion. Kaplan-Meier analysis showed an association with disease recurrence (P=0.03). Concurrently, IHC staining for AR found significantly lower levels of AR expression within those tumors deleted for PTEN (P<0.05). To validate these observations we interrogated a copy number alteration and gene expression profiling dataset of 64 PCa samples, 17 of which were PTEN deleted. We confirmed the predictive value of PTEN deletion in disease recurrence (P=0.03). PTEN deletion was also linked to diminished expression of PTEN (P<0.01) and AR (P=0.02). Furthermore, gene set enrichment analysis revealed a diminished expression of genes downstream of AR signalling in PTEN deleted tumors.
Conclusions: Altogether, our data suggest that PTEN deleted tumors expressing low levels of AR may represent a worse prognostic subset of PCa establishing a challenge for therapeutic management.
Figures
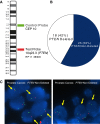
Figure 1
Dual color FISH analysis of PTEN deletion in primary PCa. A) BAC DNA mapping to chromosome 10q23.3 (PTEN) was fluorescently labelled and co-hybridized with fluorescent Centromere 10 control probe to detect PTEN deletion in tumor samples. B) PTEN deletion status of 43 primary PCa samples determined by FISH. C) FISH for PTEN status in representative interphase nuclei of prostate samples. On the left panel, the FISH image shows 1 red signal (10q23.3 locus) and two green signals (centromere 10) per nuclei indicating a PTEN deletion. On the right panel, the FISH image shows two red signals and two green signals in the nuclei indicating no PTEN deletion.
Figure 2
Prognostic value of PTEN deletion in PCa. Kaplan-Meier recurrence-free survival analysis based on PTEN deletion status determined by FISH (n=43). _P_-value (log-rank test) indicated.
Figure 3
AR IHC staining of PCa. Examples of nuclear AR staining corresponding to the variation of the adjusted H-score scale with A=10, B=28, C=71 and D=76. Original magnification 400X.
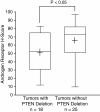
Figure 4
PTEN deletion is associated with low AR expression in PCa. Adjusted H-score of nuclear AR (IHC) was compared between PCa with and without PTEN deletion determined by FISH. The box-plot shows the mean (+ sign), the 25th, 50th (median), 75th percentiles of AR H-score including the minimum and maximum (two-sided Mann–Whitney _U_-test, _P_-value indicated).
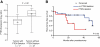
Figure 5
PTEN deletion predicts disease recurrence in an independent PCa cohort. A) In the dataset of Lapointe et al., PTEN deletion status of 64 PCa as determined by array CGH was associated with low PTEN mRNA levels measured by gene expression profiling. The box-plot shows the mean (+ sign), the 25th, 50th (median), 75th percentiles of PTEN expression including the minimum and maximum (unequal variance _t_-test, _P_-Value indicated). Values are reported as log2 ratios, normalized to the sample-set mean. B) Kaplan-Meier analysis of recurrence-free survival based on PTEN deletion status of a subset of the PCa cohort for which the clinical follow-up was available (n=29). _P_-value (log-rank test) indicated.
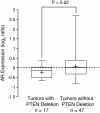
Figure 6
PTEN deletion is associated with low AR expression in an independent PCa cohort.PTEN deletion status as determined by array CGH was associated with low AR mRNA levels as measured by gene expression profiling of 64 PCa cases from the dataset of Lapointe et al. The box-plot shows the mean (+ sign), the 25th, 50th (median), 75th percentiles of AR expression including the minimum and maximum (unequal variance _t_-test, _P_-value indicated). Values are reported as log2 ratios, normalized to the sample-set mean.
Figure 7
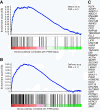
PTEN status is associated with AR signalling. GSEA was performed with previously published gene expression data of 64 prostate tumors (Lapointe et al.) stratified by their PTEN genomic status. Two androgen responsive gene sets were tested: A) a curated set of 71 genes (NELSON_RESPONSE_TO_ANDROGEN_UP, Nelson et al.) from the Molecular Signatures database (MSigDB, C2) and B) a set of 204 genes reported by DePrimo et al. GSEA identified enrichment of androgen responsive genes in PTEN positive samples. The enrichment score (ES, y-axis) reflects the degree to which an androgen responsive gene set is overrepresented at the top ranked list of genes according to the PTEN status (ranked in descending order from left to right, x-axis). Enrichment is evidenced by the early positive deflection of the running sum curve (blue line). A thousand permutations were done and the false discovery rate estimated (FDR). C) Genes from Nelson et al. that contribute to the enrichment core.
Similar articles
- PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer.
Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA. Sircar K, et al. J Pathol. 2009 Aug;218(4):505-13. doi: 10.1002/path.2559. J Pathol. 2009. PMID: 19402094 - Interaction of the Androgen Receptor, ETV1, and PTEN Pathways in Mouse Prostate Varies with Pathological Stage and Predicts Cancer Progression.
Higgins J, Brogley M, Palanisamy N, Mehra R, Ittmann MM, Li JZ, Tomlins SA, Robins DM. Higgins J, et al. Horm Cancer. 2015 Jun;6(2-3):67-86. doi: 10.1007/s12672-014-0215-9. Epub 2015 Jan 29. Horm Cancer. 2015. PMID: 25631336 Free PMC article. - Molecular profiling of ETS and non-ETS aberrations in prostate cancer patients from northern India.
Ateeq B, Kunju LP, Carskadon SL, Pandey SK, Singh G, Pradeep I, Tandon V, Singhai A, Goel A, Amit S, Agarwal A, Dinda AK, Seth A, Tsodikov A, Chinnaiyan AM, Palanisamy N. Ateeq B, et al. Prostate. 2015 Jul 1;75(10):1051-62. doi: 10.1002/pros.22989. Epub 2015 Mar 23. Prostate. 2015. PMID: 25809148 Free PMC article. - Interplay Among PI3K/AKT, PTEN/FOXO and AR Signaling in Prostate Cancer.
Yan Y, Huang H. Yan Y, et al. Adv Exp Med Biol. 2019;1210:319-331. doi: 10.1007/978-3-030-32656-2_14. Adv Exp Med Biol. 2019. PMID: 31900915 Review. - Genomic Rearrangements of PTEN in Prostate Cancer.
Phin S, Moore MW, Cotter PD. Phin S, et al. Front Oncol. 2013 Sep 17;3:240. doi: 10.3389/fonc.2013.00240. Front Oncol. 2013. PMID: 24062990 Free PMC article. Review.
Cited by
- Race and prostate cancer: genomic landscape.
Arenas-Gallo C, Owiredu J, Weinstein I, Lewicki P, Basourakos SP, Vince R Jr, Al Hussein Al Awamlh B, Schumacher FR, Spratt DE, Barbieri CE, Shoag JE. Arenas-Gallo C, et al. Nat Rev Urol. 2022 Sep;19(9):547-561. doi: 10.1038/s41585-022-00622-0. Epub 2022 Aug 9. Nat Rev Urol. 2022. PMID: 35945369 Review. - The role of phosphoinositide-regulated actin reorganization in chemotaxis and cell migration.
Wu CY, Lin MW, Wu DC, Huang YB, Huang HT, Chen CL. Wu CY, et al. Br J Pharmacol. 2014 Dec;171(24):5541-54. doi: 10.1111/bph.12777. Epub 2014 Nov 24. Br J Pharmacol. 2014. PMID: 25420930 Free PMC article. Review. - Quality of Life and Sexual Health in the Aging of PCa Survivors.
Gacci M, Baldi E, Tamburrino L, Detti B, Livi L, De Nunzio C, Tubaro A, Gravas S, Carini M, Serni S. Gacci M, et al. Int J Endocrinol. 2014;2014:470592. doi: 10.1155/2014/470592. Epub 2014 Mar 17. Int J Endocrinol. 2014. PMID: 24744780 Free PMC article. Review. - Copy number alterations are associated with metastatic-lethal progression in prostate cancer.
Wang X, Grasso CS, Jordahl KM, Kolb S, Nyame YA, Wright JL, Ostrander EA, Troyer DA, Lance R, Feng Z, Dai JY, Stanford JL. Wang X, et al. Prostate Cancer Prostatic Dis. 2020 Sep;23(3):494-506. doi: 10.1038/s41391-020-0212-8. Epub 2020 Feb 18. Prostate Cancer Prostatic Dis. 2020. PMID: 32071439 Free PMC article. - DNA alterations in the tumor genome and their associations with clinical outcome in prostate cancer.
Liu W. Liu W. Asian J Androl. 2016 Jul-Aug;18(4):533-42. doi: 10.4103/1008-682X.177120. Asian J Androl. 2016. PMID: 26975494 Free PMC article. Review.
References
- Canadian Cancer Society’s Steering Committee on Cancer Statistics. Canadian Cancer Statistics 2011. Toronto: Canadian Cancer Society; 2011.
- Petrylak DP, Tangen CM, Hussain MH, Lara PN Jr, Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M. et al.Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous