Identification of TP53 as an acute lymphocytic leukemia susceptibility gene through exome sequencing - PubMed (original) (raw)
Case Reports
Identification of TP53 as an acute lymphocytic leukemia susceptibility gene through exome sequencing
Bradford C Powell et al. Pediatr Blood Cancer. 2013 Jun.
Abstract
Although acute lymphocytic leukemia (ALL) is the most common childhood cancer, genetic predisposition to ALL remains poorly understood. Whole-exome sequencing was performed in an extended kindred in which five individuals had been diagnosed with leukemia. Analysis revealed a nonsense variant of TP53 which has been previously reported in families with sarcomas and other typical Li Fraumeni syndrome-associated cancers but never in a familial leukemia kindred. This unexpected finding enabled identification of an appropriate sibling bone marrow donor and illustrates that exome sequencing will reveal atypical clinical presentations of even well-studied genes.
Copyright © 2012 Wiley Periodicals, Inc.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
Dr. Richard Gibbs receives compensation as a consultant for GE Healthcare. Donna Muzny has been a co-owner of SeqWright, which also held investments in Lasergen and Visigen. She divested from SeqWright and Visigen in 2012. The authors declare no other potential conflict of interest.
Figures
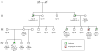
Figure 1
Clinical pedigree of the individuals involved in this study. Arrows indicate consultands who presented for genetic evaluation and “dx.” indicates age at cancer diagnosis. This kindred includes five individuals who had developed leukemia with only one other individual being affected with esophageal carcinoma. Exome sequence analysis was performed the affected individuals III-7 (patient 1) and IV-2 (patient 2) as well as the familial control (mother of patient 1; individual II-5) who did not transmit the cancer risk. Targeted analysis for the TP53 familial mutation by Sanger sequencing subsequently revealed that individual IV-4 carries the mutation (E+(TP53)) but individuals IV-1 and IV-3 do not (E−(TP53)).
Figure 2
The heterozygous TP53 mutation c.916C>T was present in the two affected individuals but not in the nontransmitting parent. (A) Germline variants were filtered by familial relationships, rarity (allele frequency <= 0.01 in 1000 Genomes Project data and NIEHS Environmental Genome Project) and potential for functional change to protein sequence (missense, nonsense, frameshift, codon insertion/deletion or splice-altering). 231 rare variants (72 predicted to change protein sequence) were present in both affected individuals but not in the familial control. Further details regarding these variants are given in Supplementary Table 1. (B) The variant c.916C>T was present in two patients but not in the familial control through whole exome sequencing. Sequence reads near this mutation are displayed using the Integrative Genomics Viewer, with reads colored by strand (red and blue for positive and negative strand, respectively). The view is centered at cDNA position 916 of TP53 and variant (non-reference) base calls are highlighted for each subject.
Similar articles
- Case report: TP53 c.848G>A germline mutation as a possible screening target at initial diagnosis for acute lymphoblastic leukemia.
Hua F, Hu Y, He GC, Lai SH, He Y, Zhang S, Deng Y, Han Y, Liu XD, Yang K, Zhong HX, Xiao J, Zheng ZZ, Yi H. Hua F, et al. Hematology. 2024 Dec;29(1):2377860. doi: 10.1080/16078454.2024.2377860. Epub 2024 Jul 15. Hematology. 2024. PMID: 39007733 - Whole-exome analysis of a Li-Fraumeni family trio with a novel TP53 PRD mutation and anticipation profile.
Franceschi S, Spugnesi L, Aretini P, Lessi F, Scarpitta R, Galli A, Congregati C, Caligo MA, Mazzanti CM. Franceschi S, et al. Carcinogenesis. 2017 Sep 1;38(9):938-943. doi: 10.1093/carcin/bgx069. Carcinogenesis. 2017. PMID: 28911001 - Mutational analysis of TP53 gene in Tunisian familial hematological malignancies and sporadic acute leukemia cases.
Hamadou WS, Besbes S, Bourdon V, Youssef YB, Laatiri MA, Noguchi T, Khélif A, Sobol H, Soua Z. Hamadou WS, et al. Fam Cancer. 2017 Jan;16(1):153-157. doi: 10.1007/s10689-016-9931-3. Fam Cancer. 2017. PMID: 27619989 - TP53 Mutations in Hypodiploid Acute Lymphoblastic Leukemia.
Comeaux EQ, Mullighan CG. Comeaux EQ, et al. Cold Spring Harb Perspect Med. 2017 Mar 1;7(3):a026286. doi: 10.1101/cshperspect.a026286. Cold Spring Harb Perspect Med. 2017. PMID: 28003275 Free PMC article. Review. - Hematologic malignancies and Li-Fraumeni syndrome.
Swaminathan M, Bannon SA, Routbort M, Naqvi K, Kadia TM, Takahashi K, Alvarado Y, Ravandi-Kashani F, Patel KP, Champlin R, Kantarjian H, Strong L, DiNardo CD. Swaminathan M, et al. Cold Spring Harb Mol Case Stud. 2019 Feb 1;5(1):a003210. doi: 10.1101/mcs.a003210. Print 2019 Feb. Cold Spring Harb Mol Case Stud. 2019. PMID: 30709875 Free PMC article. Review.
Cited by
- From regulation to deregulation of p53 in hematologic malignancies: implications for diagnosis, prognosis and therapy.
Ahmadi SE, Rahimian E, Rahimi S, Zarandi B, Bahraini M, Soleymani M, Safdari SM, Shabannezhad A, Jaafari N, Safa M. Ahmadi SE, et al. Biomark Res. 2024 Nov 14;12(1):137. doi: 10.1186/s40364-024-00676-9. Biomark Res. 2024. PMID: 39538363 Free PMC article. Review. - TP53 variants underlying pediatric low-hypodiploidy B-cell acute lymphoblastic leukemia demonstrate diverse origins and may persist as a hematopoietic clone in remission.
Itov A, Ilyasova K, Soldatkina O, Kazakova A, Kozeev V, Semchenkova A, Osipova E, Boichenko E, Volchkov E, Popov A, Zerkalenkova E, Roumiantseva J, Novichkova G, Karachunskiy A, Olshanskaya Y. Itov A, et al. EJHaem. 2024 Aug 20;5(5):1010-1013. doi: 10.1002/jha2.986. eCollection 2024 Oct. EJHaem. 2024. PMID: 39415918 Free PMC article. - The Hidden Variable: A Case of Dasatinib-Induced Respiratory Failure.
Drekolias D, Gadela NV, Syeda A, Jacob J. Drekolias D, et al. Cureus. 2020 Dec 4;12(12):e11892. doi: 10.7759/cureus.11892. Cureus. 2020. PMID: 33415044 Free PMC article. - Germ-Line TP53 Mutation in an Adolescent With CMML/Atypical CML and Familiar Cancer Predisposition.
Nucera S, Fazio G, Piazza R, Rigamonti S, Fontana D, Gambacorti Passerini C, Maitz S, Rovelli A, Biondi A, Cazzaniga G, Balduzzi A. Nucera S, et al. Hemasphere. 2020 Sep 17;4(5):e460. doi: 10.1097/HS9.0000000000000460. eCollection 2020 Oct. Hemasphere. 2020. PMID: 33163904 Free PMC article. - A Systematic Literature Review of Whole Exome and Genome Sequencing Population Studies of Genetic Susceptibility to Cancer.
Rotunno M, Barajas R, Clyne M, Hoover E, Simonds NI, Lam TK, Mechanic LE, Goldstein AM, Gillanders EM. Rotunno M, et al. Cancer Epidemiol Biomarkers Prev. 2020 Aug;29(8):1519-1534. doi: 10.1158/1055-9965.EPI-19-1551. Epub 2020 May 28. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 32467344 Free PMC article.
References
- Weintraub M, Lin AY, Franklin J, et al. Absence of germline p53 mutations in familial lymphoma. Oncogene. 1996;12:687–91. - PubMed
- Pötzsch C, Schaefer HE, Lübbert M. Familial and metachronous malignant lymphoma: absence of constitutional p53 mutations. American Journal of Hematology. 1999;62:144–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous