A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus - PubMed (original) (raw)
Review
A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus
Nicki Tiffin et al. Orphanet J Rare Dis. 2013.
Abstract
Systemic lupus erythematosus (SLE) is a chronic systemic autoimmune disease with variable clinical presentation frequently affecting the skin, joints, haemopoietic system, kidneys, lungs and central nervous system. It can be life threatening when major organs are involved. The full pathological and genetic mechanisms of this complex disease are yet to be elucidated; although roles have been described for environmental triggers such as sunlight, drugs and chemicals, and infectious agents. Cellular processes such as inefficient clearing of apoptotic DNA fragments and generation of autoantibodies have been implicated in disease progression. A diverse array of disease-associated genes and microRNA regulatory molecules that are dysregulated through polymorphism and copy number variation have also been identified; and an effect of ethnicity on susceptibility has been described.
Figures
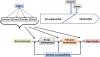
Figure 1
Stages in the pathogenesis of SLE. Environmental triggers (hormones, viruses etc.) and genetic factors along with other chance events, act on the immune system to initiate autoimmunity. Symptoms of clinical illness appear soon after pathogenic autoimmunity develops.
Figure 2
General hypothesis for the pathogenesis of SLE. Increased production of apoptotic blebs and/or reduced clearance of apoptotic blebs lead to the release of chromatin into the circulation. Presence of chromatin in circulation leads to the activation of antigen-presenting cells (APCs) and the formation of pathogenic immune complexes that incite glomerulonephritis. From Munoz et al. 2008 [20], copyright © 2011 by SAGE Publishing, reprinted by permission of SAGE.
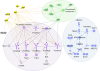
Figure 3
Network analysis of known SLE-associated genes. Network analysis using Ingenuity Pathway Analysis software shows regulatory interactions between almost half of the known SLE-associated genes, shown as shaded molecules. Three sub-networks are circled. Potential regulatory partners that participate in the networks but are not previously associated with SLE are shown as non-shaded molecules. Several IgG molecules have multiple interactions with network members, shown as dotted lines.
Similar articles
- The Epigenetics of Lupus Erythematosus.
Wu H, Chang C, Lu Q. Wu H, et al. Adv Exp Med Biol. 2020;1253:185-207. doi: 10.1007/978-981-15-3449-2_7. Adv Exp Med Biol. 2020. PMID: 32445096 Review. - Low copy numbers of complement C4 and homozygous deficiency of C4A may predispose to severe disease and earlier disease onset in patients with systemic lupus erythematosus.
Jüptner M, Flachsbart F, Caliebe A, Lieb W, Schreiber S, Zeuner R, Franke A, Schröder JO. Jüptner M, et al. Lupus. 2018 Apr;27(4):600-609. doi: 10.1177/0961203317735187. Epub 2017 Oct 19. Lupus. 2018. PMID: 29050534 Free PMC article. - SiLEncing SLE: the power and promise of small noncoding RNAs.
Rigby RJ, Vinuesa CG. Rigby RJ, et al. Curr Opin Rheumatol. 2008 Sep;20(5):526-31. doi: 10.1097/BOR.0b013e328304b45e. Curr Opin Rheumatol. 2008. PMID: 18698172 Review. - Role of microRNA-15a in autoantibody production in interferon-augmented murine model of lupus.
Yuan Y, Kasar S, Underbayev C, Vollenweider D, Salerno E, Kotenko SV, Raveche E. Yuan Y, et al. Mol Immunol. 2012 Sep;52(2):61-70. doi: 10.1016/j.molimm.2012.04.007. Epub 2012 May 11. Mol Immunol. 2012. PMID: 22578383 Free PMC article. - MicroRNAs in the key events of systemic lupus erythematosus pathogenesis.
Husakova M. Husakova M. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016 Sep;160(3):327-42. doi: 10.5507/bp.2016.004. Epub 2016 Mar 21. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016. PMID: 27003314 Review.
Cited by
- Effects of Major Epigenetic Factors on Systemic Lupus Erythematosus.
Farivar S, Shaabanpour Aghamaleki F. Farivar S, et al. Iran Biomed J. 2018 Sep;22(5):294-302. doi: 10.29252/ibj.22.5.294. Epub 2018 May 27. Iran Biomed J. 2018. PMID: 29803202 Free PMC article. Review. - Central pathways causing fatigue in neuro-inflammatory and autoimmune illnesses.
Morris G, Berk M, Walder K, Maes M. Morris G, et al. BMC Med. 2015 Feb 6;13:28. doi: 10.1186/s12916-014-0259-2. BMC Med. 2015. PMID: 25856766 Free PMC article. Review. - The Neuro-Immune Pathophysiology of Central and Peripheral Fatigue in Systemic Immune-Inflammatory and Neuro-Immune Diseases.
Morris G, Berk M, Galecki P, Walder K, Maes M. Morris G, et al. Mol Neurobiol. 2016 Mar;53(2):1195-1219. doi: 10.1007/s12035-015-9090-9. Epub 2015 Jan 20. Mol Neurobiol. 2016. PMID: 25598355 Review. - Association of microRNA-34a rs2666433 (A/G) Variant with Systemic Lupus Erythematosus in Female Patients: A Case-Control Study.
Ismail NM, Toraih EA, Mohammad MHS, Alshammari EM, Fawzy MS. Ismail NM, et al. J Clin Med. 2021 Oct 30;10(21):5095. doi: 10.3390/jcm10215095. J Clin Med. 2021. PMID: 34768615 Free PMC article. - APRIL gene polymorphism and serum sAPRIL levels in children with systemic lupus erythematosus.
Namazi S, Tajik N, Ziaee V, Sadr M, Soltani S, Rezaei A, Zoghi S, Rezaei N. Namazi S, et al. Clin Rheumatol. 2017 Apr;36(4):831-836. doi: 10.1007/s10067-016-3466-8. Epub 2016 Nov 23. Clin Rheumatol. 2017. PMID: 27878683
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical