Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS - PubMed (original) (raw)
. 2013 Feb 20;77(4):639-46.
doi: 10.1016/j.neuron.2013.02.004. Epub 2013 Feb 12.
Kevin F Bieniek, Tania F Gendron, Thomas Caulfield, Wen-Lang Lin, Mariely Dejesus-Hernandez, Marka M van Blitterswijk, Karen Jansen-West, Joseph W Paul 3rd, Rosa Rademakers, Kevin B Boylan, Dennis W Dickson, Leonard Petrucelli
Affiliations
- PMID: 23415312
- PMCID: PMC3593233
- DOI: 10.1016/j.neuron.2013.02.004
Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS
Peter E A Ash et al. Neuron. 2013.
Abstract
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) are devastating neurodegenerative disorders with clinical, genetic, and neuropathological overlap. Hexanucleotide (GGGGCC) repeat expansions in a noncoding region of C9ORF72 are the major genetic cause of FTD and ALS (c9FTD/ALS). The RNA structure of GGGGCC repeats renders these transcripts susceptible to an unconventional mechanism of translation-repeat-associated non-ATG (RAN) translation. Antibodies generated against putative GGGGCC repeat RAN-translated peptides (anti-C9RANT) detected high molecular weight, insoluble material in brain homogenates, and neuronal inclusions throughout the CNS of c9FTD/ALS cases. C9RANT immunoreactivity was not found in other neurodegenerative diseases, including CAG repeat disorders, or in peripheral tissues of c9FTD/ALS. The specificity of C9RANT for c9FTD/ALS is a potential biomarker for this most common cause of FTD and ALS. These findings have significant implications for treatment strategies directed at RAN-translated peptides and their aggregation and the RNA structures necessary for their production.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1. Anti-C9RANT immunoreactivity is specific to c9FTD/ALS
(A) Schematic representation of the possible protein products generated by RAN translation of expanded GGGGCC repeats in the three alternate reading frames. (B, C) Immunoreactivity of each anti-C9RANT antibody (Rb5822 and RB5823) towards indicated peptides was measured by adsorbing peptides onto carbon electrodes in 96-well MSD plates, and co-incubating wells with anti-C9RANT and a SULFO-tagged anti-rabbit secondary antibody. Anti-C9RANT binding to respective peptides was quantified by measuring the intensity of emitted light upon electrochemical stimulation of the plate using the MSD Sector Imager 2400. The amino acid sequence for (GX)5 is Gly-Met-Gly-Asp-Gly-Ser-Gly-Leu-Gly-Thr. (D) Western blot analysis of cerebellar tissue urea fractions from C9ORF72 repeat expansion and non-expansion FTLD cases using anti-C9RANT. Note the high molecular weight product (Arrow). (E) Anti-C9RANT immunoreactivity in cerebellar urea fractions from FTLD-TDP and ALS cases with or without expanded GGGGCC repeats, as assessed by dot blot. Each dot represents one case. See also Figure S1. (F-I) Immunohistochemistry with each anti-C9RANT antibody revealed that abundant neuronal inclusion in the cerebellum of c9FTD (F, H), but not in sporadic FTLD-TDP (G, I). C9RANT-immunoreactive lesions were granular neuronal cytoplasmic inclusions (seen clearly in the Purkinje cell shown in inset of F, H). Scale Bar=50 μm in main images, 20 μm in insets.
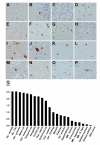
Figure 2. Regional neuropathology of C9RANT
C9RANT-immunoreactive neuronal inclusions were observed throughout the central nervous system, including cerebellum (A– cerebellar molecular layer, B–cerebellar Purkinje cell layer, C–cerebellar internal granular layer), neocortex (D–frontal cortex, E–temporal cortex, F–motor cortex), subcortical gray matter (G– amygdala, H–dentate gyrus of the hippocampus, I–CA3 of the hippocampus, J–lateral geniculate nucleus, K–lateral thalamus, L–medial geniculate nucleus, N–globus pallidus, O– hypothalamus, P–nucleus basalis of Meynert), and to a lesser extent the brainstem (M– substantia nigra). (Q) Semi-quantitative pathology scoring in these aforementioned regions, as well as in the entorhinal cortex, medulla, putamen, midbrain, and spinal cord reveals variable, C9RANT immunoreactive inclusions throughout the central nervous system. Cbl=cerebellum; ctx= cortex; Hp=hippocampus; MB=midbrain; S=substantia; SC=spinal cord; Subthal=subthalamic; Thal=thalamus.
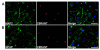
Figure 3. C9RANT-immunoreactive inclusions are present in neurons but not glia
Double-label immunofluorescence was performed on hippocampal tissue from a c9FTD case using anti-C9RANT (Rb5823) antiserum and the neuronal marker, MAP2, or the astrocytic marker, GFAP. Note that C9RANT-immunoreactive inclusions localize exclusively to neurons (A) and are not found in astrocytes (B). Scale bar = 50 μm.
Figure 4. Specificity of C9RANT pathology
p62 immunolabeling of neuronal intranuclear inclusions in c9FTD/ALS (A; cerebellum), Huntington’s disease (C; basal ganglia), spinocerebellar ataxia type 3 (E; pons), and spinal and bulbar muscular atrophy (Kennedy’s disease) (G; medulla). Anti-C9RANT-positive inclusions are specific to c9FTD/ALS (B) and absent from these other CAG repeat disorders (D–Huntington’s disease, F–spinocerebellar ataxia type 2, H–Kennedy’s disease). Additionally, C9RANT pathology is predominantly neuronal, with no inclusions in the heart (I), kidney (J), or spleen (K). The only other organ where C9RANT lesions were found was the testes, where C9RANT immunoreactive cytoplasmic and nuclear inclusions were noted in Sertoli cells (L). Scale bars in H and L=30 μm; scale bar in B and inset of L=6 μm. (See also Figure S2 and Table S1)
Comment in
- RANTing about C9orf72.
Lashley T, Hardy J, Isaacs AM. Lashley T, et al. Neuron. 2013 Feb 20;77(4):597-8. doi: 10.1016/j.neuron.2013.02.009. Neuron. 2013. PMID: 23439112 - Neurodegenerative disease: Researchers identify the protein in c9FTD/ALS inclusions.
Bible E. Bible E. Nat Rev Neurol. 2013 Apr;9(4):183. doi: 10.1038/nrneurol.2013.39. Epub 2013 Mar 12. Nat Rev Neurol. 2013. PMID: 23478465 No abstract available.
Similar articles
- Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS.
Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Gendron TF, et al. Acta Neuropathol. 2013 Dec;126(6):829-44. doi: 10.1007/s00401-013-1192-8. Epub 2013 Oct 16. Acta Neuropathol. 2013. PMID: 24129584 Free PMC article. - c9RAN translation: a potential therapeutic target for the treatment of amyotrophic lateral sclerosis and frontotemporal dementia.
Gendron TF, Cosio DM, Petrucelli L. Gendron TF, et al. Expert Opin Ther Targets. 2013 Sep;17(9):991-5. doi: 10.1517/14728222.2013.818659. Epub 2013 Jul 12. Expert Opin Ther Targets. 2013. PMID: 23844663 - Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72.
Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB, Rademakers R, Dickson DW. Murray ME, et al. Acta Neuropathol. 2011 Dec;122(6):673-90. doi: 10.1007/s00401-011-0907-y. Epub 2011 Nov 15. Acta Neuropathol. 2011. PMID: 22083254 Free PMC article. - Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion.
Babić Leko M, Župunski V, Kirincich J, Smilović D, Hortobágyi T, Hof PR, Šimić G. Babić Leko M, et al. Behav Neurol. 2019 Jan 15;2019:2909168. doi: 10.1155/2019/2909168. eCollection 2019. Behav Neurol. 2019. PMID: 30774737 Free PMC article. Review. - Insights into the pathogenic mechanisms of Chromosome 9 open reading frame 72 (C9orf72) repeat expansions.
Todd TW, Petrucelli L. Todd TW, et al. J Neurochem. 2016 Aug;138 Suppl 1:145-62. doi: 10.1111/jnc.13623. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27016280 Review.
Cited by
- Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS.
Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, Bieniek KF, DeTure M, Lee WC, Biendarra SM, Davis MD, Baker MC, Perkerson RB, van Blitterswijk M, Stetler CT, Rademakers R, Link CD, Dickson DW, Boylan KB, Li H, Petrucelli L. Prudencio M, et al. Nat Neurosci. 2015 Aug;18(8):1175-82. doi: 10.1038/nn.4065. Epub 2015 Jul 20. Nat Neurosci. 2015. PMID: 26192745 Free PMC article. - ALS-associated C21ORF2 variant disrupts DNA damage repair, mitochondrial metabolism, neuronal excitability and NEK1 levels in human motor neurons.
Zelina P, de Ruiter AA, Kolsteeg C, van Ginneken I, Vos HR, Supiot LF, Burgering BMT, Meye FJ, Veldink JH, van den Berg LH, Pasterkamp RJ. Zelina P, et al. Acta Neuropathol Commun. 2024 Sep 4;12(1):144. doi: 10.1186/s40478-024-01852-6. Acta Neuropathol Commun. 2024. PMID: 39227882 Free PMC article. - Validated assays for the quantification of C9orf72 human pathology.
Salomonsson SE, Maltos AM, Gill K, Aladesuyi Arogundade O, Brown KA, Sachdev A, Sckaff M, Lam KJK, Fisher IJ, Chouhan RS, Van Laar VS, Marley CB, McLaughlin I, Bankiewicz KS, Tsai YC, Conklin BR, Clelland CD. Salomonsson SE, et al. Sci Rep. 2024 Jan 8;14(1):828. doi: 10.1038/s41598-023-50667-3. Sci Rep. 2024. PMID: 38191789 Free PMC article. - Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes.
Busch JI, Unger TL, Jain N, Tyler Skrinak R, Charan RA, Chen-Plotkin AS. Busch JI, et al. Hum Mol Genet. 2016 Jul 1;25(13):2681-2697. doi: 10.1093/hmg/ddw127. Epub 2016 Apr 28. Hum Mol Genet. 2016. PMID: 27126638 Free PMC article. - Quantitative Nucleocytoplasmic Transport Assays in Cellular Models of Neurodegeneration.
Vanneste J, Vercruysse T, Van Damme P, Van Den Bosch L, Daelemans D. Vanneste J, et al. Bio Protoc. 2020 Jun 20;10(12):e3659. doi: 10.21769/BioProtoc.3659. eCollection 2020 Jun 20. Bio Protoc. 2020. PMID: 33659329 Free PMC article.
References
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 NS074121-01/NS/NINDS NIH HHS/United States
- P50 NS72187/NS/NINDS NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- R21 NS074121/NS/NINDS NIH HHS/United States
- R01 NS063964/NS/NINDS NIH HHS/United States
- P50 NS072187/NS/NINDS NIH HHS/United States
- R01 ES20395/ES/NIEHS NIH HHS/United States
- R01 ES020395/ES/NIEHS NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- R01 NS080882/NS/NINDS NIH HHS/United States
- R01 AG026251/AG/NIA NIH HHS/United States
- R01 NS077402/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous