MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients - PubMed (original) (raw)
MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients
Kai Doberstein et al. Neoplasia. 2013 Feb.
Abstract
A disintegrin and metalloproteinase 17 (ADAM17) is a metalloprotease that is overexpressed in many cancer types, including renal cancers. However, the regulatory mechanisms of ADAM17 in cancer development and progression are poorly understood. In the present work, we provide evidence using overexpression and inhibition of microRNA 145 (miR-145) that miR-145 negatively regulates ADAM17 expression. Furthermore, we show that ADAM17 negatively regulates miR-145 through tumor necrosis factor-α, resulting in a reciprocal negative feedback loop. In this study, the expression of ADAM17 and miR-145 correlated negatively in renal cancer tumor tissues and cell lines, suggesting an important regulatory mechanism. Additionally, we showed that the regulation of ADAM17 is partly involved in the effects of miR-145 on proliferation and migration, whereas no involvement in chemosensitivity was observed. Importantly, in the healthy kidney, miR-145 was detected in different cell types including tubular cells, which are considered the origin of renal cancer. In renal cancer cell lines, miR-145 expression was strongly suppressed by methylation. In summary, miR-145 is downregulated in renal cancer patients, which leads to the up-regulation of ADAM17 in renal cancer. Importantly, miR-145 and ADAM17 are regulated in a reciprocal negative feedback loop.
Figures
Figure 1
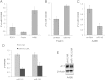
miR-145 regulates ADAM17 expression in renal cancer cell line. (A) Predicted duplex formation between human miR-145 (hsamiR-145; top) and human ADAM17 3′UTR (bottom). (B) Sequence of the miR-145 binding site within the ADAM17 3′UTR of human (Hsa), chimpanzee (Ptr), Rhesus macaque (Mml), mouse (Mmu), and rat (Rno). (C) Relative real-time PCR of miR-145 expression in relation to U48 expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (D) Western blot analysis for ADAM17 (A17) of total lysates of HRCepiC, RCC4, 786-0, A498, and Foehn cells. β-Actin antibody was used as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. (E) Relative real-time PCR of ADAM17 mRNA expression in relation to β-actin expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (F) Real-time PCR of miR-145 expression relative to U48 expression in RCC tissue compared to normal renal tissue of 15 renal cancer patients (bright gray bars) and real-time PCR of ADAM17 expression in RCC tumor samples compared to normal renal tissue of the same patients (dark gray bars). The values were evaluated with the Δ-Δ _C_T method. Correlations were calculated with Student's t test, normal tissue against tumor tissue for miR-145 and ADAM17. (G) Plot of fold change of miR-145 related to fold change of ADAM17 for each patient; r, Pearson correlation coefficient; a, regression line. (H) Plot of fold change of miR-145 related to fold change of c-Myc for each patient. (I) Plot of fold change of miR-145 related to fold change of PAI-1 for each patient. (J) Overview of a tissue section of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemistry with an aquaporin-1 (red)-specific antibody. Scale bar represents 300 _µ_m. (K) Enlarged picture of J showing the miR-145-expressing blood vessel. Scale bar represents 300 _µ_m. (L) Enlarged picture of J. Scale bar represents 300 _µ_m. (M) Enlarged picture of I showing miR-145-expressing (blue) and aquaporin-1 (red)-expressing tubular cells. Scale bar represents 50 _µ_m. (N) Section of a glomerulus of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemical analysis for aquaporin-1 (red). Scale bar represents 50 _µ_m.
Figure 1
miR-145 regulates ADAM17 expression in renal cancer cell line. (A) Predicted duplex formation between human miR-145 (hsamiR-145; top) and human ADAM17 3′UTR (bottom). (B) Sequence of the miR-145 binding site within the ADAM17 3′UTR of human (Hsa), chimpanzee (Ptr), Rhesus macaque (Mml), mouse (Mmu), and rat (Rno). (C) Relative real-time PCR of miR-145 expression in relation to U48 expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (D) Western blot analysis for ADAM17 (A17) of total lysates of HRCepiC, RCC4, 786-0, A498, and Foehn cells. β-Actin antibody was used as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. (E) Relative real-time PCR of ADAM17 mRNA expression in relation to β-actin expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (F) Real-time PCR of miR-145 expression relative to U48 expression in RCC tissue compared to normal renal tissue of 15 renal cancer patients (bright gray bars) and real-time PCR of ADAM17 expression in RCC tumor samples compared to normal renal tissue of the same patients (dark gray bars). The values were evaluated with the Δ-Δ _C_T method. Correlations were calculated with Student's t test, normal tissue against tumor tissue for miR-145 and ADAM17. (G) Plot of fold change of miR-145 related to fold change of ADAM17 for each patient; r, Pearson correlation coefficient; a, regression line. (H) Plot of fold change of miR-145 related to fold change of c-Myc for each patient. (I) Plot of fold change of miR-145 related to fold change of PAI-1 for each patient. (J) Overview of a tissue section of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemistry with an aquaporin-1 (red)-specific antibody. Scale bar represents 300 _µ_m. (K) Enlarged picture of J showing the miR-145-expressing blood vessel. Scale bar represents 300 _µ_m. (L) Enlarged picture of J. Scale bar represents 300 _µ_m. (M) Enlarged picture of I showing miR-145-expressing (blue) and aquaporin-1 (red)-expressing tubular cells. Scale bar represents 50 _µ_m. (N) Section of a glomerulus of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemical analysis for aquaporin-1 (red). Scale bar represents 50 _µ_m.
Figure 1
miR-145 regulates ADAM17 expression in renal cancer cell line. (A) Predicted duplex formation between human miR-145 (hsamiR-145; top) and human ADAM17 3′UTR (bottom). (B) Sequence of the miR-145 binding site within the ADAM17 3′UTR of human (Hsa), chimpanzee (Ptr), Rhesus macaque (Mml), mouse (Mmu), and rat (Rno). (C) Relative real-time PCR of miR-145 expression in relation to U48 expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (D) Western blot analysis for ADAM17 (A17) of total lysates of HRCepiC, RCC4, 786-0, A498, and Foehn cells. β-Actin antibody was used as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. (E) Relative real-time PCR of ADAM17 mRNA expression in relation to β-actin expression in HRCepiC, RCC4, 786-0, A498, and Foehn cells. (F) Real-time PCR of miR-145 expression relative to U48 expression in RCC tissue compared to normal renal tissue of 15 renal cancer patients (bright gray bars) and real-time PCR of ADAM17 expression in RCC tumor samples compared to normal renal tissue of the same patients (dark gray bars). The values were evaluated with the Δ-Δ _C_T method. Correlations were calculated with Student's t test, normal tissue against tumor tissue for miR-145 and ADAM17. (G) Plot of fold change of miR-145 related to fold change of ADAM17 for each patient; r, Pearson correlation coefficient; a, regression line. (H) Plot of fold change of miR-145 related to fold change of c-Myc for each patient. (I) Plot of fold change of miR-145 related to fold change of PAI-1 for each patient. (J) Overview of a tissue section of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemistry with an aquaporin-1 (red)-specific antibody. Scale bar represents 300 _µ_m. (K) Enlarged picture of J showing the miR-145-expressing blood vessel. Scale bar represents 300 _µ_m. (L) Enlarged picture of J. Scale bar represents 300 _µ_m. (M) Enlarged picture of I showing miR-145-expressing (blue) and aquaporin-1 (red)-expressing tubular cells. Scale bar represents 50 _µ_m. (N) Section of a glomerulus of a normal kidney stained by in situ hybridization for miR-145 (blue) and by immunohistochemical analysis for aquaporin-1 (red). Scale bar represents 50 _µ_m.
Figure 2
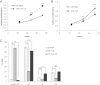
miR-145 binds directly to the 3′UTR of ADAM17 mRNA. (A) Western blot analysis for ADAM17 of total lysates of Foehn cells transfected with control RNA (ctrl RNA), miR-145 mimic (miR-145), miR-145 inhibitor (miR-145 inh), or specific siRNA against ADAM17 (A17-siRNA). β-Actin was used to determine equal loading. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted (n = 4). (B) Western blot analysis for ADAM17 of total lysates of A498 cells transfected with control RNA (ctrl RNA), miR-145 mimic (miR-145), miR-145 inhibitor (miR-145 inh), or specific siRNA against ADAM17 (A17-siRNA). β-Actin was used to determine equal loading. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted (n = 3). (C) Luciferase assays with reporter constructs containing either ADAM17 3′UTR with the predicted miR-145 binding site or the mutated binding site. Furthermore, a reporter construct carrying the complement sequence of the mature form of miR-145 was transfected as positive control. The Renilla luciferase activity for each construct was normalized with firefly luciferase activities. Fold change = (_S_renilla/_S_firefly)/(_C_renilla/_C_firefly).
Figure 3
Inhibition of miR-143 reduces ADAM17 expression. (A) Relative real-time PCR of miR-143 expression in relation to U48 expression in RCC4, Foehn, and A498 cells. (B) Relative real-time PCR of miR-143 expression in relation to U48 expression in Foehn cells that were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). (C) Relative real-time PCR of miR-143 expression in relation to U48 expression in A498 cells that were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). (D) Relative real-time PCR of miR-143 and miR-145 expression in relation to U48 expression in A498 cells that were transfected with control LNA (ctrl LNA) or miR-143 LNA. (E) Western blot analysis for ADAM17 (A17) of total lysates of D. β-Actin antibody was used as a loading control.
Figure 4
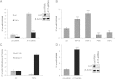
miR-145 reduces proliferation by induction of apoptosis. (A) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (n = 3). (B) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after siRNA transfection using an MTT assay (n = 3). (C) Foehn cells were transfected with control RNA (ctrl RNA), miR-145 mimic (miR-145), or miR-145 inhibitor (miR-145 inh). Cell cycle analysis was performed as described in Materials and Methods section. The percentages of Foehn cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase are depicted (n = 3). (D) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Forty-eight hours after transfection, 10 _µ_g/ml cisplatin (CDDP) was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed in a graph (n = 3). (E) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Forty-eight hours after transfection, 10 _µ_g/ml CDDP was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed (n = 3). (F) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or miR-145 inhibitor with ADAM17 siRNA (miR-145 inh + A17si). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using an MTT assay. (G) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Invasion assay was performed using matrigelcoated transwell chambers. (H) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). The percentage of cells in the sub-G1/G0 (apoptosis) phase is displayed in a graph. (I) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Cell lysates were investigated by Western blot using antibodies against ADAM17 (A17) and β-actin as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. *P < .05, **P < .01, and P < .001 are considered statistically significant. ns, not significant.
Figure 4
miR-145 reduces proliferation by induction of apoptosis. (A) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (n = 3). (B) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after siRNA transfection using an MTT assay (n = 3). (C) Foehn cells were transfected with control RNA (ctrl RNA), miR-145 mimic (miR-145), or miR-145 inhibitor (miR-145 inh). Cell cycle analysis was performed as described in Materials and Methods section. The percentages of Foehn cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase are depicted (n = 3). (D) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Forty-eight hours after transfection, 10 _µ_g/ml cisplatin (CDDP) was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed in a graph (n = 3). (E) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Forty-eight hours after transfection, 10 _µ_g/ml CDDP was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed (n = 3). (F) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or miR-145 inhibitor with ADAM17 siRNA (miR-145 inh + A17si). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using an MTT assay. (G) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Invasion assay was performed using matrigelcoated transwell chambers. (H) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). The percentage of cells in the sub-G1/G0 (apoptosis) phase is displayed in a graph. (I) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Cell lysates were investigated by Western blot using antibodies against ADAM17 (A17) and β-actin as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. *P < .05, **P < .01, and P < .001 are considered statistically significant. ns, not significant.
Figure 4
miR-145 reduces proliferation by induction of apoptosis. (A) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (n = 3). (B) Foehn cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after siRNA transfection using an MTT assay (n = 3). (C) Foehn cells were transfected with control RNA (ctrl RNA), miR-145 mimic (miR-145), or miR-145 inhibitor (miR-145 inh). Cell cycle analysis was performed as described in Materials and Methods section. The percentages of Foehn cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase are depicted (n = 3). (D) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 mimic (miR-145). Forty-eight hours after transfection, 10 _µ_g/ml cisplatin (CDDP) was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed in a graph (n = 3). (E) RCC4 cells were transfected with control RNA (ctrl RNA) or miR-145 inhibitor (miR-145 inh). Forty-eight hours after transfection, 10 _µ_g/ml CDDP was added to the cells for 24 hours. Cells were harvested and measured by cell cycle analysis as described in Materials and Methods section. The percentage of cells in the sub-G1/G0 (apoptosis), G1 (haploid genome), S (DNA synthesis), or G2 (diploid genome) phase is displayed (n = 3). (F) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or miR-145 inhibitor with ADAM17 siRNA (miR-145 inh + A17si). Twenty-four hours after RNA transfection, anchorage-dependent cell growth was measured at the time points 24, 48, and 72 hours after RNA transfection using an MTT assay. (G) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Invasion assay was performed using matrigelcoated transwell chambers. (H) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). The percentage of cells in the sub-G1/G0 (apoptosis) phase is displayed in a graph. (I) RCC4 cells were transfected with control RNA (ctrl RNA), miR-145 inhibitor (miR-145 inh), or ADAM17 siRNA together with miR-145 inhibitor (miR-145 inh + A17si). Cell lysates were investigated by Western blot using antibodies against ADAM17 (A17) and β-actin as a loading control. The densitometrical analysis of the ADAM17 expression to β-actin is additionally depicted. *P < .05, **P < .01, and P < .001 are considered statistically significant. ns, not significant.
Figure 5
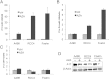
ADAM17 regulates miR-145 expression by cleavage of TNF-α. (A) Real-time PCR of miR-145 relative to U48 expression in RCC4 cells after transfection with either control RNA (ctrl RNA) or specific ADAM17 siRNA (A17-siRNA). Forty-eight hours after transfection, the cells were treated with control vehicle (bright bars) or with 10 nM TNF-α (dark bars). The Western blot analysis for ADAM17 (A17) of total lysates to control the efficient down-regulation is additionally depicted. β-Actin antibody was used as a loading control. (B) Real-time PCR of miR-145 expression relative to U48 expression in RCC4 cells after treatment for 24 hours with either control vehicle (ctrl), PMA (100 ng/ml), TNF-α (10 nM), or the metalloprotease inhibitors TAPI-0 and TAPI-2 (both 10 _µ_M). *P < .05, **P < .01, and ***P < .001 are considered statistically significant. (C) Real-time PCR of miR-145 expression relative to U48 expression (gray bars) and ADAM17 expression to β-actin expression (black bars) in RCC4 cells after treatment for 24 hours with either control vehicle (ctrl) or TNF-α (10 nM). (D) Real-time PCR of miR-145 expression relative to U48 expression in Foehn cells after transfection with either control RNA (ctrl RNA, bright bars) or specific ADAM17 siRNA (A17-siRNA, dark bars). The Western blot analysis for ADAM17 (A17) of total lysates to control the efficient down-regulation is additionally depicted. β-Actin antibody was used as a loading control.
Figure 6
miR-145 is downregulated by methylation in renal cancer cells. (A) Real-time PCR of miR-145 expression relative to U48 expression in A498, RCC4, and Foehn cells after treatment with either control vehicle (ctrl, bright bars) or AZA (10 _µ_M, dark bars) for 72 hours. (B) Real-time PCR of pri-miR-145 expression relative to β-actin expression in A498, RCC4, and Foehn cells after treatment with either control vehicle (ctrl, bright bars) or AZA (10 _µ_M, dark bars) for 72 hours. (C) Real-time PCR of miR-143 expression relative to U48 expression in A498, RCC4, and Foehn cells after treatment with either control vehicle (ctrl, bright bars) or AZA (10 _µ_M, dark bars) for 72 hours. (D) A498, RCC4, and Foehn cells after treatment with either control vehicle (-) or AZA (10 _µ_M, +) for 72 hours. Cell lysates were investigated by Western blot analysis with antibodies against ADAM17 and β-actin as a loading control.
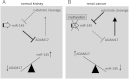
Figure 7
Reciprocal negative feedback loop of miR-145 and ADAM17 is transmitted through TNF-α. (A) Endogenous expression of miR-145 in normal kidney cells leads to the down-regulation of ADAM17 expression. As a consequence, less substrates are released by ADAM17, leading to an even higher expression of miR-145. As a result, ADAM17 is weakly expressed, whereas the miR-145 expression is high. (B) miR-145 expression is suppressed in renal cancer cells by an initial event, like promoter methylation. This leads to an up-regulation of ADAM17 expression, resulting in an increase of ADAM17-mediated cleavage of substrates like TNF-α. The released substrates then further suppress the expression of miR-145, leading to a reciprocal negative feedback loop. As a result, ADAM17 expression is elevated in renal cancer patients, whereas miR-145 is downregulated.
Similar articles
- MiR-145, a microRNA targeting ADAM17, inhibits the invasion and migration of nasopharyngeal carcinoma cells.
Wu J, Yin L, Jiang N, Guo WJ, Gu JJ, Chen M, Xia YY, Wu JZ, Chen D, Wu JF, Wang DJ, Zong D, Zhang N, Ding K, Huang T, He X. Wu J, et al. Exp Cell Res. 2015 Nov 1;338(2):232-8. doi: 10.1016/j.yexcr.2015.08.006. Epub 2015 Aug 20. Exp Cell Res. 2015. PMID: 26297956 - MiR-338-3p inhibits the proliferation and migration of gastric cancer cells by targeting ADAM17.
Chen JT, Yao KH, Hua L, Zhang LP, Wang CY, Zhang JJ. Chen JT, et al. Int J Clin Exp Pathol. 2015 Sep 1;8(9):10922-8. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26617808 Free PMC article. - MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma.
Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. Tsai WC, et al. Hepatology. 2009 May;49(5):1571-82. doi: 10.1002/hep.22806. Hepatology. 2009. PMID: 19296470 - MicroRNA-152 targets ADAM17 to suppress NSCLC progression.
Su Y, Wang Y, Zhou H, Lei L, Xu L. Su Y, et al. FEBS Lett. 2014 May 21;588(10):1983-8. doi: 10.1016/j.febslet.2014.04.022. Epub 2014 Apr 26. FEBS Lett. 2014. PMID: 24780186 - The role of ADAM17 in tumorigenesis and progression of breast cancer.
Shen H, Li L, Zhou S, Yu D, Yang S, Chen X, Wang D, Zhong S, Zhao J, Tang J. Shen H, et al. Tumour Biol. 2016 Dec;37:15359–15370. doi: 10.1007/s13277-016-5418-y. Epub 2016 Sep 22. Tumour Biol. 2016. PMID: 27658778 Review.
Cited by
- The Microrna-143/145 Cluster in Tumors: A Matter of Where and When.
Poli V, Seclì L, Avalle L. Poli V, et al. Cancers (Basel). 2020 Mar 17;12(3):708. doi: 10.3390/cancers12030708. Cancers (Basel). 2020. PMID: 32192092 Free PMC article. Review. - Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway.
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang Z, Wang X, Lin Y, Mao Y, Chen H, Luo J, Liu B, Zheng X, Xie L. Xu X, et al. Mol Cancer. 2014 May 17;13:109. doi: 10.1186/1476-4598-13-109. Mol Cancer. 2014. PMID: 24886554 Free PMC article. - MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways.
Cui SY, Wang R, Chen LB. Cui SY, et al. J Cell Mol Med. 2014 Oct;18(10):1913-26. doi: 10.1111/jcmm.12358. Epub 2014 Aug 15. J Cell Mol Med. 2014. PMID: 25124875 Free PMC article. Review. - HDAC-Linked "Proliferative" miRNA Expression Pattern in Pancreatic Neuroendocrine Tumors.
Klieser E, Urbas R, Swierczynski S, Stättner S, Primavesi F, Jäger T, Mayr C, Kiesslich T, Fazio PD, Helm K, Neureiter D. Klieser E, et al. Int J Mol Sci. 2018 Sep 15;19(9):2781. doi: 10.3390/ijms19092781. Int J Mol Sci. 2018. PMID: 30223590 Free PMC article. - miR-145 Alleviates Smooth Muscle Cell Phenotype Transition via ADAM17-Mediated ACE2 Shedding.
Wen J, Tang B, Guo L, Chen W, Tang X. Wen J, et al. Int J Hypertens. 2023 Jul 20;2023:9497716. doi: 10.1155/2023/9497716. eCollection 2023. Int J Hypertens. 2023. PMID: 37521117 Free PMC article.
References
- McLaughlin JK, Lipworth L. Epidemiologic aspects of renal cell cancer. Semin Oncol. 2000;27:115–123. - PubMed
- Campbell SC, Flanigan RC, Clark JI. Nephrectomy in metastatic renal cell carcinoma. Curr Treat Options Oncol. 2003;4:363–372. - PubMed
- Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H, Rocken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol. 2005;131:41–48. - PubMed
- Blanchot-Jossic F, Jarry A, Masson D, Bach-Ngohou K, Paineau J, Denis MG, Laboisse CL, Mosnier JF. Up-regulated expression of ADAM17 in human colon carcinoma: co-expression with EGFR in neoplastic and endothelial cells. J Pathol. 2005;207:156–163. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous