Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes - PubMed (original) (raw)
Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes
Anthony Levasseur et al. Biotechnol Biofuels. 2013.
Abstract
Background: Since its inception, the carbohydrate-active enzymes database (CAZy; http://www.cazy.org) has described the families of enzymes that cleave or build complex carbohydrates, namely the glycoside hydrolases (GH), the polysaccharide lyases (PL), the carbohydrate esterases (CE), the glycosyltransferases (GT) and their appended non-catalytic carbohydrate-binding modules (CBM). The recent discovery that members of families CBM33 and family GH61 are in fact lytic polysaccharide monooxygenases (LPMO), demands a reclassification of these families into a suitable category.
Results: Because lignin is invariably found together with polysaccharides in the plant cell wall and because lignin fragments are likely to act in concert with (LPMO), we have decided to join the families of lignin degradation enzymes to the LPMO families and launch a new CAZy class that we name "Auxiliary Activities" in order to accommodate a range of enzyme mechanisms and substrates related to lignocellulose conversion. Comparative analyses of these auxiliary activities in 41 fungal genomes reveal a pertinent division of several fungal groups and subgroups combining their phylogenetic origin and their nutritional mode (white vs. brown rot).
Conclusions: The new class introduced in the CAZy database extends the traditional CAZy families, and provides a better coverage of the full extent of the lignocellulose breakdown machinery.
Figures
Figure 1
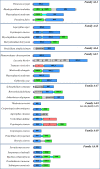
Examples of modular AA proteins. The additional modules represented include: carbohydrate-binding modules (CBMs; green), conserved modules of unknown function (X; red) and other domains (labelled grey boxes). Signal peptides (purple), proline/serine-rich linkers (cyan) and unassigned connecting regions (unlabelled grey boxes) are also shown.
Figure 2
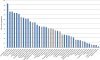
Number of AA proteins in the 41 selected fungal genomes. Basidiomycetes are depicted in blue and ascomycetes are depicted in grey.
Figure 3
Distribution of AA proteins in the 41 selected fungal genomes.
Figure 4
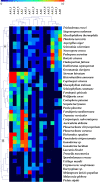
Comparison of the ligninolytic AA repertoires identified in the selected fungal genomes using double hierarchical clustering. Top tree: family number according to the AA classification. Left tree: Fungal genomes analyzed. A and B correspond to the Ascomycotina and Basidiomycotina divisions, respectively. The abundance of the different AA proteins within a family is represented by a colour scale from 0 (dark blue) to ≥ 10 occurrences (red) per species. Note that only ligninolytic AA families were selected (AA1 to AA8).
Similar articles
- Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay.
Hori C, Gaskell J, Igarashi K, Samejima M, Hibbett D, Henrissat B, Cullen D. Hori C, et al. Mycologia. 2013 Nov-Dec;105(6):1412-27. doi: 10.3852/13-072. Epub 2013 Aug 9. Mycologia. 2013. PMID: 23935027 - Comparative Genomics of Rumen Butyrivibrio spp. Uncovers a Continuum of Polysaccharide-Degrading Capabilities.
Palevich N, Kelly WJ, Leahy SC, Denman S, Altermann E, Rakonjac J, Attwood GT. Palevich N, et al. Appl Environ Microbiol. 2019 Dec 13;86(1):e01993-19. doi: 10.1128/AEM.01993-19. Print 2019 Dec 13. Appl Environ Microbiol. 2019. PMID: 31653790 Free PMC article. - The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. Cantarel BL, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D233-8. doi: 10.1093/nar/gkn663. Epub 2008 Oct 5. Nucleic Acids Res. 2009. PMID: 18838391 Free PMC article. - Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation.
Andlar M, Rezić T, Marđetko N, Kracher D, Ludwig R, Šantek B. Andlar M, et al. Eng Life Sci. 2018 Jun 27;18(11):768-778. doi: 10.1002/elsc.201800039. eCollection 2018 Nov. Eng Life Sci. 2018. PMID: 32624871 Free PMC article. Review. - Chitin-Active Lytic Polysaccharide Monooxygenases.
Courtade G, Aachmann FL. Courtade G, et al. Adv Exp Med Biol. 2019;1142:115-129. doi: 10.1007/978-981-13-7318-3_6. Adv Exp Med Biol. 2019. PMID: 31102244 Review.
Cited by
- Penicillium arizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites.
Grijseels S, Nielsen JC, Randelovic M, Nielsen J, Nielsen KF, Workman M, Frisvad JC. Grijseels S, et al. Sci Rep. 2016 Oct 14;6:35112. doi: 10.1038/srep35112. Sci Rep. 2016. PMID: 27739446 Free PMC article. - The impact of the carbohydrate-binding module on how a lytic polysaccharide monooxygenase modifies cellulose fibers.
Støpamo FG, Sulaeva I, Budischowsky D, Rahikainen J, Marjamaa K, Kruus K, Potthast A, Eijsink VGH, Várnai A. Støpamo FG, et al. Biotechnol Biofuels Bioprod. 2024 Aug 24;17(1):118. doi: 10.1186/s13068-024-02564-8. Biotechnol Biofuels Bioprod. 2024. PMID: 39182111 Free PMC article. - Optimization of synergism of a recombinant auxiliary activity 9 from Chaetomium globosum with cellulase in cellulose hydrolysis.
Kim IJ, Nam KH, Yun EJ, Kim S, Youn HJ, Lee HJ, Choi IG, Kim KH. Kim IJ, et al. Appl Microbiol Biotechnol. 2015 Oct;99(20):8537-47. doi: 10.1007/s00253-015-6592-3. Epub 2015 May 5. Appl Microbiol Biotechnol. 2015. PMID: 25936375 Free PMC article. - SACCHARIS v2: Streamlining Prediction of Carbohydrate-Active Enzyme Specificities Within Large Datasets.
Fraser ASC, Low KE, Tingley JP, Reintjes G, Thomas D, Brumer H, Abbott DW. Fraser ASC, et al. Methods Mol Biol. 2024;2836:299-330. doi: 10.1007/978-1-0716-4007-4_16. Methods Mol Biol. 2024. PMID: 38995547 - Pecularities and applications of aryl-alcohol oxidases from fungi.
Urlacher VB, Koschorreck K. Urlacher VB, et al. Appl Microbiol Biotechnol. 2021 May;105(10):4111-4126. doi: 10.1007/s00253-021-11337-4. Epub 2021 May 17. Appl Microbiol Biotechnol. 2021. PMID: 33997930 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous