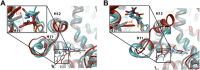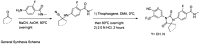Overcoming mutation-based resistance to antiandrogens with rational drug design - PubMed (original) (raw)
Overcoming mutation-based resistance to antiandrogens with rational drug design
Minna D Balbas et al. Elife. 2013.
Abstract
The second-generation antiandrogen enzalutamide was recently approved for patients with castration-resistant prostate cancer. Despite its success, the duration of response is often limited. For previous antiandrogens, one mechanism of resistance is mutation of the androgen receptor (AR). To prospectively identify AR mutations that might confer resistance to enzalutamide, we performed a reporter-based mutagenesis screen and identified a novel mutation, F876L, which converted enzalutamide into an AR agonist. Ectopic expression of AR F876L rescued the growth inhibition of enzalutamide treatment. Molecular dynamics simulations performed on antiandrogen-AR complexes suggested a mechanism by which the F876L substitution alleviates antagonism through repositioning of the coactivator recruiting helix 12. This model then provided the rationale for a focused chemical screen which, based on existing antiandrogen scaffolds, identified three novel compounds that effectively antagonized AR F876L (and AR WT) to suppress the growth of prostate cancer cells resistant to enzalutamide. DOI:http://dx.doi.org/10.7554/eLife.00499.001.
Keywords: Human; Mouse; androgen receptor; drug resistance; prostate cancer.
Conflict of interest statement
MDB, Co-inventor of patent applications covering the AR mutation and novel chemical matter described in this manuscript.
MJE, Co-inventor of a patent application covering the novel chemical matter described in this manuscript.
DJH, Co-inventor of a patent application covering the novel chemical matter described in this manuscript.
GLG, Co-inventor of a patent application covering the novel chemical matter described in this manuscript.
YS, Co-inventor of a patent application covering the novel chemical matter described in this manuscript.
PAW, Co-inventor of a patent application covering the AR mutation described in this manuscript.
CLS, Senior editor, eLife; co-inventor of enzalutamide and ARN-509; co-founder of Aragon Pharmaceuticals; co-inventor of patent applications covering the AR mutation and novel chemical matter described in this manuscript.
The other authors declare that no competing interests exist.
Figures
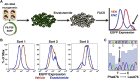
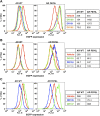
Figure 1.. Mutagenesis screen for enzalutamide resistance identifies novel AR mutation.
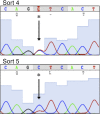
(A) A cartoon of the AR mutagenesis screen developed to identify enzalutamide resistance mutations. Briefly, cells were cotransduced with a randomly mutagenized AR cDNA library (AR*) and EGFP reporter of AR activity (Pb.PSE.EGFP), treated with 1 μM enzalutamide, and EGFP-positive cells were sorted using FACS. AR was PCR amplified and sequenced to identify relevant mutations. (B) Representative FACS histograms showing the progressive enrichment of an EGFP-positive subpopulation of LNCaP/AR*/Pb.PSE.EGFP cells post multiple rounds of enzalutamide treatment and cell sorting. (C) A Sanger sequencing trace of exon 8 within AR on the exogenous AR allele from LNCaP/AR*/Pb.PSE.EGFP cells after the fifth FACS sort. The position of the mutation is highlighted with an arrow. The alignment was performed against AR WT. AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.003
Figure 1—figure supplement 1.. Enrichment of AR W741C mutant expressing LNCaP-Pb.PSE.EGFP cells after bicalutamide treatment and EGFP sorting.
Genomic DNA was isolated from LNCaP-Pb.PSE.EGFP cells ectopically expressing either wild-type (WT) AR or mutant AR W741C, or different ratios of mutant-to-WT, and quantitative PCR was performed to test the sensitivity of the W741C-specific primers. With starting ratios of 1:100 and 1:1000 mutant-to-WT, we treated these cell mixtures with 1 μM bicalutamide for 4 days, and then FACS-sorted those that maintained/induced EGFP expression. Sorted cells were expanded and the brief bicalutamide treatment and FACS-sorting was repeated (four rounds). Genomic DNA was isolated from the sorted cell populations, and quantitative PCR was performed to test for enrichment of the W741C mutant cells. AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.004
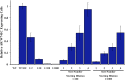
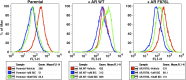
Figure 1—figure supplement 2.. Endogenous AR target gene, PSA is induced by enzalutamide in FACS-sorted cells.
Parental LNCaP-Pb.PSE.EGFP cells, and those overexpressing AR WT, the random AR mutant library (Mut Lib), and cells after each sort were treated with 1 μM enzalutamide for 24 hr in media containing full serum. RNA was then collected, reverse transcribed, and quantitative PCR performed for AR target gene KLK3 (PSA). AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.005
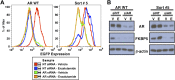
Figure 1—figure supplement 3.. Expression of EGFP and endogenous AR target gene FKBP5 remain AR-dependent in FACS-sorted cells.
LNCaP-Pb.PSE.EGFP cells overexpressing AR WT and cells from the fifth sort of our screen were transfected with 10 nM of either a non-targeting siRNA (siNT) or a siRNA against AR (siAR). They were also treated with either vehicle (V) or 1 μM enzalutamide (E). After 4 days of enzalutamide treatment and siRNA knockdown, cells were collected for both (A) flow cytometric analysis of EGFP expression and (B) western blot analysis of the AR target gene FKBP5, and to ensure we achieved good AR knockdown. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.006
Figure 1—figure supplement 4.. AR F876L mutation accounts for 50% of AR in cells after fourth sort, and further enriched after the fifth sort.
We PCR amplified AR from our LNCaP/AR*/Pb.PSE.EGFP cells after four rounds of enzalutamide treatment and FACS-sorting, and Sanger sequenced the PCR product. AR F876L (T → C) accounts for approximately 50% of the AR in these cells. This mutation is further enriched after the fifth sort, and accounts for approximately 80% of AR in that population of cells. AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.007
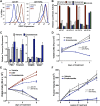
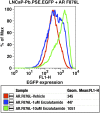
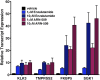
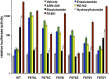
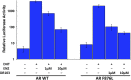
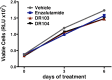
Figure 2.. AR F876L mutation converts enzalutamide into an agonist and rescues enzalutamide-induced growth inhibition.
(A) A representative FACS histogram shows the induction of AR-dependent EGFP expression by enzalutamide in LNCaP-Pb.PSE.EGFP cells ectopically expressing AR F876L. The magnitude of induction by enzalutamide (10 μM) is comparable to that conferred by the endogenous androgen DHT (1 nM). Enzalutamide treatment of LNCaP-Pb.PSE.EGFP cells ectopically expressing AR WT effectively suppressed EGFP expression. Geometric-mean fluorescence intensity for WT treated cells: vehicle (348), enzalutamide (66.4), DHT (1554); for F876L cells: vehicle (345), enzalutamide (1051), DHT (1699). (B) Cotransduction of CV1 cells with an AR-regulated firefly luciferase construct, a constitutive Renilla luciferase construct, and one of the indicated AR constructs, recapitulates the pharmacology observed in the EGFP reporter system. These cells were treated with vehicle (DMSO), antiandrogens (1 μM), or the synthetic androgen R1881 (1 nM). A dual luciferase assay was conducted on cell lysates, the firefly signal was normalized to the constitutive Renilla activity, and the data are reported as relative light units (RLUs). Notably, the bisaryl-thiohydantoin antiandrogens (enzalutamide and ARN-509) effectively induce AR F876L transcriptional activity, while structurally discrete antiandrogens (hydroxyflutamide and bicalutamide) do not impact AR F876L activity in this assay. As expected, the transcriptional activity of AR W741C or AR T877A was induced by bicalutamide or hydroxyflutamide, respectively. (C) Quantitative reverse transcription–polymerase chain reaction analysis of LNCaP/AR F876L cells shows that enzalutamide (1 μM) can induce the expression of canonical AR-regulated gene products (i.e., PSA, TMPRSS2, SGK1, and FKBP5). Relative gene expression post therapy for LNCaP/AR WT cells is included as positive controls. FL = AR F876L, data are normalized to GAPDH and represented as mean ± SD, n = 3. (D) Cell proliferation data shows that overexpression of AR F876L in a human prostate cancer cell line sensitive to enzalutamide therapy can rescue cell growth. VCaP cells overexpressing either AR WT (solid lines) or AR F876L (dashed lines) were cultured in media containing full serum, treated with either vehicle (DMSO) or 10 μM enzalutamide, and the viable cell fraction was determined at the indicated time points (data is represented as mean ± SD, n = 3). (E) Cellular proliferation data shows that enzalutamide also rescues the growth of VCaP cells expressing AR F876L in androgen-depleted media. VCaP cells overexpressing either AR WT (solid lines) or AR F876L (dashed lines) were treated with vehicle (DMSO), 1 nM DHT, or 10 μM enzalutamide, and the viable cell fraction was determined at the indicated time points (mean ± SD, n = 3). (F) A time to progression study for mice bearing subcutaneous LNCaP/AR-WT (solid lines) or LNCaP/AR-F876L (dashed lines) xenografts further highlights the genotype-dependent pharmacology of enzalutamide. Inoculated animals were treated once daily through oral gavage with either vehicle or enzalutamide (30 mg/kg), and tumor size was monitored weekly (11–16 tumors per treatment group). While enzalutamide potently suppressed the growth of LNCaP/AR-WT tumors, LNCaP/AR-F876L tumors exposed to enzalutamide grew with kinetics roughly equivalent to either vehicle treatment arm. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.008
Figure 2—figure supplement 1.. Dose-dependent induction of EGFP expression by enzalutamide in LNCaP-Pb.PSE.EGFP cells expressing AR F876L.
LNCaP-Pb.PSE.EGFP cells ectopically expressing AR F876L were treated with vehicle, 1 or 10 μM enzalutamide for 4 days. Cells were then collected for FACS-analysis of EGFP expression. Geometric-mean fluorescence intensity is indicated in the table. AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.009
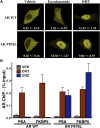
Figure 2—figure supplement 2.. Enzalutamide induces AR F876L nuclear translocation and DNA binding to AR enhancer elements.
(A) LNCaP cells were transfected with EYFP-tagged wild-type AR or AR F876L in androgen depleted media containing vehicle, 1 μM enzalutamide, or 1 nM DHT. Representative confocal images are shown. Average nuclear-to-cytoplasmic ratios for EYFP are displayed (±SD, n = 3). (B) LNCaP cells stably overexpressing either AR WT or AR F876L were cultured in androgen-depleted media for 4 days, then treated with vehicle (VEH), 10 μM enzalutamide (ENZ), or 1 nM DHT for 4 hr. AR chromatin immuoprecipitation was performed, and real-time PCR quantification of PSA enhancer and FKBP5 enhancer is shown (percent input mean ± SD, n = 3). AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.010
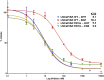
Figure 2—figure supplement 3.. Enzalutamide binds to AR F876L with higher affinity than to AR WT.
Representative competition binding curves, showing displacement of 18F-FDHT in LNCaP/AR WT and LNCaP/AR F876L cells by increasing concentrations of cold DHT or enzalutamide (ENZ). The median inhibitory concentration (IC50) values from this experiment are displayed (error bars represent the SD of triplicate measurements). AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.011
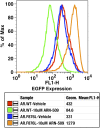
Figure 2—figure supplement 4.. AR F876L mutation converts ARN-509 into an AR agonist.
LNCaP-Pb.PSE.EGFP cells ectopically expressing either AR WT or AR F876L were treated with vehicle or 10 μM ARN-509. After 4 days of treatment, cells were collected for analysis of EGFP expression (FL1-H), geometric-mean fluorescence is shown in the table below. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.012
Figure 2—figure supplement 5.. Dose-dependent agonism in AR F876L expressing cells treated with enzalutamide or ARN-509.
LNCaP cells ectopically expressing AR F876L were cultured in androgen-depleted media (10% CSS) for 48 hr, then treated with the indicated dose of antiandrogen for 24 hr, and qRT-PCR was performed to assess the expression of the indicated AR target gene. AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.013
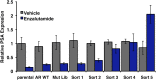
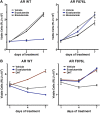
Figure 2—figure supplement 6.. Ectopic expression of AR F876L in CWR22Pc cells confers resistance to enzalutamide and rescues growth in androgen-depleted media.
(A) CWR22Pc cells stably expressing either AR WT or AR F876L were plated in full serum media containing vehicle, 1 μM enzalutamide, or 10 μM bicalutamide. CellTiterGLO assay was performed on days 1, 4, and 7 to measure cell viability (mean relative light units [RLU] ± SD, n = 3). (B) CWR22Pc cells stably expressing either AR WT or AR F876L were plated in full serum media containing vehicle, 1 μM enzalutamide, or 0.1 nM DHT. CellTiterGLO assay was performed on days 1, 4, and 7 to measure cell viability (mean relative light units [RLU] ± SD, n = 3). AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.014
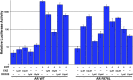
Figure 2—figure supplement 7.. Other amino acid substitutions at Phe876 modify the pharmacology of second-generation antiandrogens.
ARE(4X)-luciferase assay for additional F876 substitutions. CV1 cells were cotransfected with an ARE(4X)-firefly luciferase construct, SV40 Renilla luciferase construct, and one of the designated AR constructs. The cells were treated with 10 μM of the indicated antiandrogens, and a dual luciferase assay was performed on the lysates, and normalized to Renilla luciferase (mean ± SD, n = 3). AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.015
Figure 2—figure supplement 8.. Bicalutamide is a modest inhibitor of AR F876L transcriptional activity.
Parental LNCaP-Pb.PSE.EGFP cells and those transduced with AR WT or AR F876L were treated with vehicle, 1 or 10 μM bicalutamide (BIC) for 4 days, and then collected for flow cytometric analysis of EGFP expression (FL1-H). Geometric-mean fluorescence intensity of EGFP is displayed in the table below each histogram plot. DOI:
http://dx.doi.org/10.7554/eLife.00499.016
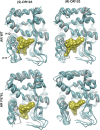
Figure 3.. Molecular dynamics simulations predict a novel binding mode for bisaryl-thiohydantoin antiandrogens and the basis for agonism toward AR F876L.
(A) Structures of the antiandrogens bicalutamide (top), enzalutamide (middle), and ARN-509 (bottom) oriented to highlight the common and discrete regions of the molecules. The A-D annotation of the rings is indicated. (B) A magnified view of the cocrystal structure of AR W741L (gray) and bicalutamide (gold) shows the antiandrogen's spatial relationship to the H11 and H12 pockets and residue F876 (blue). In this agonist conformation, the C-ring of bicalutamide does not interact with F876. (C) A magnified view of the initial-docked models of enzalutamide (gold) and ARN-509 (cyan) calculated using coordinates from 2AXA in which residue 741 is a tryptophan. The model suggests that the loss of torsional freedom imposed by the thiohydantoin B-ring imposes conformational restrictions on the antagonists that force the C-ring toward F876 and the ‘H11 pocket'. (D) The lowest-energy 10-ns MD models for enzalutamide with AR WT (red) and AR F876L (cyan) overlaid on 1Z95 (gray—agonist reference structure). The F876L mutation allows for cooperative changes in neighboring residues which, when bound to enzalutamide, enable H12 to adopt a more agonist-like conformation. (E) An analogous view of the lowest-energy 10-ns MD models for ARN-509 with AR WT (red) and AR F876L (cyan) overlaid on 1Z95 (gray) shows a similar effect for F876L on the positioning of H11 and H12. These simulations also point to the comparatively larger dislocation in H12 by ARN-509 in AR WT, presumably owing to favorable steric interactions between the spirocyclobutyl ring and H12. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.019
Figure 3—figure supplement 1.. Overlay of predicted enzalutamide or ARN-509 simulations and solved agonist crystal structure.
The coordinates for three 10-ns simulations with the indicated receptor (cyan) and drug compound (gold) were overlaid on the 1Z95 structure (gray) to highlight structural differences between the agonist conformation of the AR W741L/bicalutamide complex. Note the evidence of H12 dislocation in AR WT/antiandrogen complexes that is less evident for AR F876L. Bicalutamide was deleted for visual clarity. For the simulations conducted for the enzalutamide/AR F876L complex, one suspected outlier (red) was detected among the initial three MD simulations. Five additional simulations were conducted, and they recapitulated the majority tendency observed for the initial MD simulations. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.020
Figure 3—figure supplement 2.. A zoomed in view of the H11 pocket from the enzalutamide and ARN-509 MD simulations.
The lowest-energy 10-ns MD models for enzalutamide (A) and ARN-509 (B) with AR WT (red) and AR F876L (cyan) overlaid on 1Z95 (gray—agonist reference structure), with an inset showing a zoomed in view of the region around residue 876, that includes distances between close hydrophobic atoms on each receptor:ligand pair. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.021
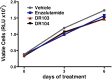
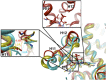
Figure 4.. A focused chemical screen identifies novel antagonists of AR F876L.
(A) A tabular summary of the bioactivity of the novel antiandrogens in the LNCaP/AR/Pb.PSE.EGFP cell-based assay shows the importance of a carefully designed D-ring for competent inhibition of AR F876L. Antagonism is indicated with a ‘−’ symbol, and agonism is indicated with a ‘+’ sign. The asterisk is situated over the shared carbon atom in position 4 that joins the bisaryl-thiohydantoin scaffold to the respective ‘substituent’. The source data is outlined in Figure 4—figure supplement 1. (B) A proliferation assay for VCaP prostate cancer cells overexpressing either wild-type AR (left) or AR F876L (right) shows that (±)-DR103 effectively inhibits the growth of both models, while enzalutamide and the close structural analogue DR101 only inhibit the growth of VCaP/AR WT. Data are reported as mean ± SD, n = 3. (C) A view of the lowest-energy conformations of enzalutamide (cyan), ARN-509 (gold), and (S)-DR103 (red) in complex with AR F876L highlights the greater dislocation of H12 and the loop between H11 and H12 uniquely conferred by (S)-DR103. The color scheme invoked for AR F876L matches the respective antiandrogen. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.022
Figure 4—figure supplement 1.. EGFP reporter assay for AR activity with DR series compounds.
LNCaP-Pb.PSE.EGFP cells ectopically expressing either AR WT or AR F876L were treated with vehicle (DMSO) or 10 μM of the indicated DR-series compound. After 4 days of treatment, cells were collected and FACS analysis for EGFP expression was performed. Geometric-mean fluorescence intensity is indicated in the adjacent table. DOI:
http://dx.doi.org/10.7554/eLife.00499.023
Figure 4—figure supplement 2.. (±)-DR103 effectively competes with DHT for AR binding and induction of AR-regulated luciferase.
CV1 cells were transfected with AR WT or AR F876L, 4XARE-luciferase, and SV40 renilla luciferase expression constructs. They were cultured in androgen-depleted media supplemented with the indicated androgen/antiandrogen for 36 hr, and a dual-luciferase assay was performed on cell lysates. Luciferase activity is represented by relative light units (RLU) ± SD, n = 3. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.024
Figure 4—figure supplement 3.. (±)-DR103 is a more potent antagonist for AR F876L than AR WT.
CV1 cells were transfected with AR WT or AR F876L, 4XARE-luciferase, and SV40 renilla luciferase expression constructs. They were cultured in androgen-depleted media supplemented with the indicated androgen/antiandrogens for 36 hr and a dual-luciferase assay was performed on cell lysates. DHT concentration was 1 nM. Luciferase activity is represented by relative light units (RLU) ± SD, n = 3. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.025
Figure 4—figure supplement 4.. Growth inhibition of CWR22Pc cells overexpressing AR WT or AR F876L with (±)-DR103 treatment.
CWR22Pc cells ectopically expressing wild-type AR or AR F876L, cultured in full-serum-containing media, were treated with vehicle (DMSO) or 10 µM of enzalutamide or DR103. CellTiterGLO assay was performed on days 1, 4, and 7 to determine cell viability (relative light units [RLU] ± SD, n = 3). AR: androgen receptor. DOI:
http://dx.doi.org/10.7554/eLife.00499.026
Figure 4—figure supplement 5.. Novel antiandrogen, (±)-DR103, efficiently inhibits AR signaling and induces PARP cleavage in cells expressing both AR WT and AR F876L.
VCaP cells ectopically expressing either AR WT or AR F876L were treated for 4 days with vehicle or 10 µM of the indicated antiandrogen (VEH = vehicle, DMSO; ENZ = enzalutamide) in media containing 10% FBS. Whole-cell lysates were analyzed by western blot. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.027
Figure 4—figure supplement 6.. DU145 cells treated with (±)-DR103 and DR104 display no significant growth inhibition.
DU145 cells were cultured in full-serum-containing media with 10 μM of the indicated antiandrogens. CellTiterGLO assay was performed on days 0, 3, and 6 to determine cell viability (relative light units [RLU] ± SD, n = 3). DOI:
http://dx.doi.org/10.7554/eLife.00499.028
Figure 4—figure supplement 7.. Overlay of predicted DR103 simulations and solved agonist crystal structure.
An overlay of three 10-ns MD simulations for (S)-DR103 docked either in AR WT or AR F876L shows the dislocation of H12 in space compared to 1Z95 (gray), consistent with the pharmacological model predicted by previous MD simulations for enzalutamide and ARN-509 in AR WT. The predicted conformations of the AR variants are highlighted in cyan, and the respective conformations of (S)- and (R)-DR103 are represented in gold. AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.029
Figure 4—figure supplement 8.. A zoomed in view of the H11 and H12 pockets of the MD simulations for AR F876L.
A magnified view of the lowest energy conformations of enzalutamide (cyan), ARN-509 (gold), and (S)-DR103 (red) in complex with AR F876L, showing the structural framework and interactions in close proximity to the antagonist C and D-rings (with distances measured). AR: androgen receptor; WT: wild-type. DOI:
http://dx.doi.org/10.7554/eLife.00499.030
General synthesis schema.
DOI:
http://dx.doi.org/10.7554/eLife.00499.031
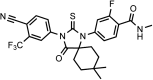
Chemical structure 1.. DR100, 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-4-oxo-2-thioxo-1,3-diazaspiro[4.4]nonan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.032
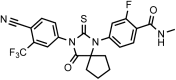
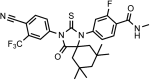
Chemical structure 2.. DR101, 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-4-oxo-2-thioxo-1,3-diazaspiro[4.5]decan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.033
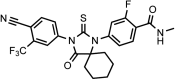
Chemical structure 3.. DR102, 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-4-oxo-2-thioxo-1,3-diazaspiro[4.6]undecan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.034
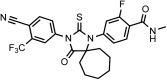
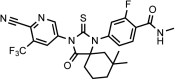
Chemical structure 4.. (±)-DR103, 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-7,7-dimethyl-4-oxo-2-thioxo-1,3-diazaspiro[4.5]decan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.035
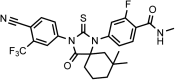
Chemical structure 5.. DR104 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-8,8-dimethyl-4-oxo-2-thioxo-1,3-diazaspiro[4.5]decan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.036
Chemical structure 6.. DR105 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-7,7,9,9-tetramethyl-4-oxo-2-thioxo-1,3-diazaspiro[4.5]decan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.037
Chemical structure 7.. (±)-DR106 4-(3-(6-cyano-5-(trifluoromethyl)pyridin-3-yl)-7,7-dimethyl-4-oxo-2-thioxo-1,3-diazaspiro[4.5]decan-1-yl)-2-fluoro-N-methylbenzamide.
DOI:
http://dx.doi.org/10.7554/eLife.00499.038
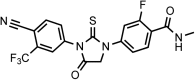
Chemical structure 8.. DR107 4-(3-(4-cyano-3-(trifluoromethyl)phenyl)-4-oxo-2-thioxoimidazolidin-1-yl)-2-fluoro-N-methylbenzamide
DOI:
http://dx.doi.org/10.7554/eLife.00499.039
Comment in
- Designer antiandrogens join the race against drug resistance.
Josan JS, Katzenellenbogen JA. Josan JS, et al. Elife. 2013 Apr 9;2:e00692. doi: 10.7554/eLife.00692. Elife. 2013. PMID: 23580166 Free PMC article. - Therapeutic resistance: two steps ahead.
Seton-Rogers S. Seton-Rogers S. Nat Rev Cancer. 2013 Jun;13(6):382-3. doi: 10.1038/nrc3532. Epub 2013 May 10. Nat Rev Cancer. 2013. PMID: 23660783 No abstract available.
Similar articles
- A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509.
Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, Moon M, Maneval EC, Chen I, Darimont B, Hager JH. Joseph JD, et al. Cancer Discov. 2013 Sep;3(9):1020-9. doi: 10.1158/2159-8290.CD-13-0226. Epub 2013 Jun 18. Cancer Discov. 2013. PMID: 23779130 - An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide).
Korpal M, Korn JM, Gao X, Rakiec DP, Ruddy DA, Doshi S, Yuan J, Kovats SG, Kim S, Cooke VG, Monahan JE, Stegmeier F, Roberts TM, Sellers WR, Zhou W, Zhu P. Korpal M, et al. Cancer Discov. 2013 Sep;3(9):1030-43. doi: 10.1158/2159-8290.CD-13-0142. Epub 2013 Jul 10. Cancer Discov. 2013. PMID: 23842682 - Molecular Dynamics Studies on the Enzalutamide Resistance Mechanisms Induced by Androgen Receptor Mutations.
Liu H, Wang L, Tian J, Li J, Liu H. Liu H, et al. J Cell Biochem. 2017 Sep;118(9):2792-2801. doi: 10.1002/jcb.25928. Epub 2017 Apr 27. J Cell Biochem. 2017. PMID: 28181296 - Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level.
Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Karantanos T, et al. Eur Urol. 2015 Mar;67(3):470-9. doi: 10.1016/j.eururo.2014.09.049. Epub 2014 Oct 8. Eur Urol. 2015. PMID: 25306226 Free PMC article. Review. - Androgen receptor-related micro RNAs in prostate cancer and their role in antiandrogen drug resistance.
Rezaei S, Mahjoubin Tehran M, Sahebkar A, Jalili A, Aghaee-Bakhtiari SH. Rezaei S, et al. J Cell Physiol. 2020 Apr;235(4):3222-3234. doi: 10.1002/jcp.29275. Epub 2019 Oct 10. J Cell Physiol. 2020. PMID: 31599460 Review.
Cited by
- Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer.
Kregel S, Chen JL, Tom W, Krishnan V, Kach J, Brechka H, Fessenden TB, Isikbay M, Paner GP, Szmulewitz RZ, Vander Griend DJ. Kregel S, et al. Oncotarget. 2016 May 3;7(18):26259-74. doi: 10.18632/oncotarget.8456. Oncotarget. 2016. PMID: 27036029 Free PMC article. - Development and exploitation of a novel mutant androgen receptor modelling strategy to identify new targets for advanced prostate cancer therapy.
O'Neill D, Jones D, Wade M, Grey J, Nakjang S, Guo W, Cork D, Davies BR, Wedge SR, Robson CN, Gaughan L. O'Neill D, et al. Oncotarget. 2015 Sep 22;6(28):26029-40. doi: 10.18632/oncotarget.4347. Oncotarget. 2015. PMID: 26267320 Free PMC article. - TLE3 loss confers AR inhibitor resistance by facilitating GR-mediated human prostate cancer cell growth.
Palit SA, Vis D, Stelloo S, Lieftink C, Prekovic S, Bekers E, Hofland I, Šuštić T, Wolters L, Beijersbergen R, Bergman AM, Győrffy B, Wessels LF, Zwart W, van der Heijden MS. Palit SA, et al. Elife. 2019 Dec 19;8:e47430. doi: 10.7554/eLife.47430. Elife. 2019. PMID: 31855178 Free PMC article. - Apalutamide: A Review in Non-Metastatic Castration-Resistant Prostate Cancer.
Al-Salama ZT. Al-Salama ZT. Drugs. 2019 Sep;79(14):1591-1598. doi: 10.1007/s40265-019-01194-x. Drugs. 2019. PMID: 31489589 Review. - A molecule inducing androgen receptor degradation and selectively targeting prostate cancer cells.
Auvin S, Öztürk H, Abaci YT, Mautino G, Meyer-Losic F, Jollivet F, Bashir T, de Thé H, Sahin U. Auvin S, et al. Life Sci Alliance. 2019 Aug 20;2(4):e201800213. doi: 10.26508/lsa.201800213. Print 2019 Aug. Life Sci Alliance. 2019. PMID: 31431473 Free PMC article.
References
- Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the Resp model. J Phys Chem. 1993;97:10269. doi: 10.1021/j100142a004. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- R25 CA096945/CA/NCI NIH HHS/United States
- K08 CA140946/CA/NCI NIH HHS/United States
- R01 CA155169/CA/NCI NIH HHS/United States
- R01 CA089489/CA/NCI NIH HHS/United States
- CA089489/CA/NCI NIH HHS/United States
- P50 CA069568/CA/NCI NIH HHS/United States
- R25-CA096945/CA/NCI NIH HHS/United States
- CA155169/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials