Resources for methylome analysis suitable for gene knockout studies of potential epigenome modifiers - PubMed (original) (raw)
Resources for methylome analysis suitable for gene knockout studies of potential epigenome modifiers
Gareth A Wilson et al. Gigascience. 2012.
Abstract
Background: Methylated DNA immunoprecipitation (MeDIP) is a popular enrichment based method and can be combined with sequencing (termed MeDIP-seq) to interrogate the methylation status of cytosines across entire genomes. However, quality control and analysis of MeDIP-seq data have remained to be a challenge.
Results: We report genome-wide DNA methylation profiles of wild type (wt) and mutant mouse cells, comprising 3 biological replicates of Thymine DNA glycosylase (Tdg) knockout (KO) embryonic stem cells (ESCs), in vitro differentiated neural precursor cells (NPCs) and embryonic fibroblasts (MEFs). The resulting 18 methylomes were analysed with MeDUSA (Methylated DNA Utility for Sequence Analysis), a novel MeDIP-seq computational analysis pipeline for the identification of differentially methylated regions (DMRs). The observed increase of hypermethylation in MEF promoter-associated CpG islands supports a previously proposed role for Tdg in the protection of regulatory regions from epigenetic silencing. Further analysis of genes and regions associated with the DMRs by gene ontology, pathway, and ChIP analyses revealed further insights into Tdg function, including an association of TDG with low-methylated distal regulatory regions.
Conclusions: We demonstrate that MeDUSA is able to detect both large-scale changes between cells from different stages of differentiation and also small but significant changes between the methylomes of cells that only differ in the KO of a single gene. These changes were validated utilising publicly available datasets and confirm TDG's function in the protection of regulatory regions from epigenetic silencing.
Figures
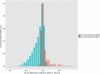
Figure 1
Comparison of MEDIPS normalised MeDIP-seq data with RRBS[25]and BS-seq[26]data. (a) ESC Tdg +/− MeDIP vs ESC wt RRBS. (b) NPC Tdg +/− MeDIP vs NPC wt RRBS. (c) MEF wt MeDIP vs MEF wt RRBS. (d) ESC Tdg +/− MeDIP vs ESC wt BS-seq. (e) NPC Tdg +/− MeDIP vs NPC wt BS-seq.
Figure 2
Mouse NPC methylomes viewed in the UCSC browser. Each sample has 3 tracks representing total coverage (red) and coverage on forward (green) and reverse (blue) strands. The data range on the Y-axis of each track represents the read depth in reads per million (RPM). Box a highlights a Tdg −/− hypomethylated DMR in exon 2 of Prdm6. Box b indicates an example of potential non-CpG methylation where the methylation signal is driven by a single strand, in this case the reverse strand.
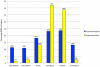
Figure 3
Number of DMRs identified in comparisons of samples from different stages of differentiation. DMRs hypermethylated in the more differentiated state are shown in blue, those hypomethylated in the more differentiated state are in yellow. (a) All DMRs. (b) DMRs located in CpG Island regions.
Figure 4
Difference in methylation score (ESCms – NPCms) calculated from BS-seq data[26]for each DMR called from the MeDIP cohorts. Regions displaying ESC hypermethylation in the BS-seq data will score >0, those displaying hypomethylation will score <0. Frequency of hypermethylated MeDIP DMRs are shown in red, hypomethylated DMRs in blue, indicating the concordance between datasets.
Figure 5
Proportion of DMRs found in different genomic features. DMRs hypermethylated in MEF Tdg −/− are shown in blue, those hypomethylated in MEF Tdg −/− are in yellow. The numbers represent the amount of DMRs found in each feature type.
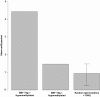
Figure 6
Overlap between MEF Tdg −/− DMRs and LMRs[26]represented by Observed/Expected ratio. Data for randomly selected genomic regions also shown, bars indicate the maximum and minimum ratio achieved from 1,000 permutations.
Similar articles
- Analyses of methylomes of upland and lowland switchgrass (Panicum virgatum) ecotypes using MeDIP-seq and BS-seq.
Dworkin M, Xie S, Saha M, Thimmapuram J, Kalavacharla VK. Dworkin M, et al. BMC Genomics. 2017 Nov 7;18(1):851. doi: 10.1186/s12864-017-4218-0. BMC Genomics. 2017. PMID: 29115936 Free PMC article. - A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation450 BeadChip(®) for methylome profiling.
Clark C, Palta P, Joyce CJ, Scott C, Grundberg E, Deloukas P, Palotie A, Coffey AJ. Clark C, et al. PLoS One. 2012;7(11):e50233. doi: 10.1371/journal.pone.0050233. Epub 2012 Nov 29. PLoS One. 2012. PMID: 23209683 Free PMC article. - DNA methylome profiling at single-base resolution through bisulfite sequencing of 5mC-immunoprecipitated DNA.
Jia Z, Shi Y, Zhang L, Ren Y, Wang T, Xing L, Zhang B, Gao G, Bu R. Jia Z, et al. BMC Biotechnol. 2018 Feb 6;18(1):7. doi: 10.1186/s12896-017-0409-7. BMC Biotechnol. 2018. PMID: 29409498 Free PMC article. Retracted. - Analyzing the cancer methylome through targeted bisulfite sequencing.
Lee EJ, Luo J, Wilson JM, Shi H. Lee EJ, et al. Cancer Lett. 2013 Nov 1;340(2):171-8. doi: 10.1016/j.canlet.2012.10.040. Epub 2012 Nov 28. Cancer Lett. 2013. PMID: 23200671 Free PMC article. Review. - Comprehensive analysis of DNA-methylation in mammalian tissues using MeDIP-chip.
Pälmke N, Santacruz D, Walter J. Pälmke N, et al. Methods. 2011 Feb;53(2):175-84. doi: 10.1016/j.ymeth.2010.07.006. Epub 2010 Jul 16. Methods. 2011. PMID: 20638478 Review.
Cited by
- Comprehensive analysis of DNA methylation data with RnBeads.
Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Assenov Y, et al. Nat Methods. 2014 Nov;11(11):1138-1140. doi: 10.1038/nmeth.3115. Epub 2014 Sep 28. Nat Methods. 2014. PMID: 25262207 Free PMC article. - DNA methylation analysis of murine hematopoietic side population cells during aging.
Taiwo O, Wilson GA, Emmett W, Morris T, Bonnet D, Schuster E, Adejumo T, Beck S, Pearce DJ. Taiwo O, et al. Epigenetics. 2013 Oct;8(10):1114-22. doi: 10.4161/epi.26017. Epub 2013 Aug 15. Epigenetics. 2013. PMID: 23949429 Free PMC article. - Role of epigenomic mechanisms in the onset and management of insulin resistance.
Izquierdo AG, Crujeiras AB. Izquierdo AG, et al. Rev Endocr Metab Disord. 2019 Mar;20(1):89-102. doi: 10.1007/s11154-019-09485-0. Rev Endocr Metab Disord. 2019. PMID: 30887406 Review. - MEDIPIPE: an automated and comprehensive pipeline for cfMeDIP-seq data quality control and analysis.
Zeng Y, Ye W, Stutheit-Zhao EY, Han M, Bratman SV, Pugh TJ, He HH. Zeng Y, et al. Bioinformatics. 2023 Jul 1;39(7):btad423. doi: 10.1093/bioinformatics/btad423. Bioinformatics. 2023. PMID: 37402621 Free PMC article. - The common bisulfite-conversion-based techniques to analyze DNA methylation in human cancers.
Jeddi F, Faghfuri E, Mehranfar S, Soozangar N. Jeddi F, et al. Cancer Cell Int. 2024 Jul 9;24(1):240. doi: 10.1186/s12935-024-03405-2. Cancer Cell Int. 2024. PMID: 38982390 Free PMC article. Review.
References
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. - DOI - PMC - PubMed
- Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials