Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner - PubMed (original) (raw)
Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner
Sunny Sun-Kin Chan et al. Cell Stem Cell. 2013.
Abstract
Mesp1 is regarded as the master regulator of cardiovascular development, initiating the cardiac transcription factor cascade to direct the generation of cardiac mesoderm. To define the early embryonic cell population that responds to Mesp1, we performed pulse inductions of gene expression over tight temporal windows following embryonic stem cell differentiation. Remarkably, instead of promoting cardiac differentiation in the initial wave of mesoderm, Mesp1 binds to the Tal1 (Scl) +40 kb enhancer and generates Flk-1+ precursors expressing Etv2 (ER71) and Tal1 that undergo hematopoietic differentiation. The second wave of mesoderm responds to Mesp1 by differentiating into PDGFRα+ precursors that undergo cardiac differentiation. Furthermore, in the absence of serum-derived factors, Mesp1 promotes skeletal myogenic differentiation. Lineage tracing revealed that the majority of yolk sac and many adult hematopoietic cells derive from Mesp1+ precursors. Thus, Mesp1 is a context-dependent determination factor, integrating the stage of differentiation and the signaling environment to specify different lineage outcomes.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
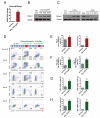
Figure 1. Opposing outcomes of early versus late induction of Mesp1
(A) Quantitative RT-PCR (n=3) and (B) immunoblot showing Mesp1 induction by Dox. (C) Immunoblot demonstrating that Mesp1 induction is tightly regulated by Dox. (D) FACS profile and (E-H) quantification of mesoderm (Flk-1/PDGFRα), hematopoietic (c-Kit/CD41, c-Kit/CD45) and cardiac markers (cTnT) during EB differentiation. Dox (500 ng/ml) was applied over 24 hr as indicated by red boxes. (E) The late 24 hr pulse of Mesp1 induction from day 3-4 increased Flk-1+ PDGFRα+ presumptive cardiac mesoderm at day 4 (left, n=15) and cTnT+ cardiomyocytes at day 8 (right, n=15). (F) The early 24 hr pulse of Mesp1 induction from day 2-3 promoted Flk-1+ PDGFRα+ presumptive early unpatterned mesoderm at day 3.25 (left, n=5) and Flk-1+ PDGFRα− presumptive lateral plate mesoderm at day 4 (right, n=10). (G,H) Mesp1 day 2-3 induction increased several hematopoietic progenitor populations at day 6, including c-Kit+ CD41+ (G, left, n=10), c-Kit+ CD45+ (G, right, n=8), c-Kit− CD41+ (H, left, n=10) and c-Kit− CD45+ (H, right, n=8). Mean ± SEM is shown in panels A, E, F, G and H. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox. See also Figure S1
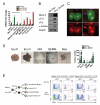
Figure 2. Characterization of differentiated cell types promoted by Mesp1 in early or late mesoderm
(A) Quantitative RT-PCR for hematopoietic- and cardiac-specific markers for day 6 EBs (n=3). Note that the early pulse (day 2-3) of Mesp1 induction (green bars) upregulated hematopoietic but not cardiac specific markers (versus No Dox, gray bars), whereas the late pulse (day 3-4, red bars) did the opposite. (B) Immunoblot demonstrating the upregulation of cardiac-specific proteins in day 8 EBs subjected to the late pulse of Mesp1 induction. (C) Immunostaining for cTnT (green) or α-actinin (green) and CX43 (red) in Mesp1- induced EB-derived cardiac cells. (D) Hematopoietic colonies induced by Mesp1 (left) and quantification (right, n=3-6). Note that the early pulse of Mesp1 induction increased the numbers of both primitive (Ery-P) and definitive (Ery-D, GM, GEMM, Mac) hematopoietic cell types. (E) Scheme for determining cell autonomy of Mesp1-induced hematopoiesis (left) and FACS analysis of hematopoietic markers (c-Kit/CD41) in day 6 EBs (right). Note that both c-Kit+ CD41+ progenitors and c-Kit− CD41+ differentiated cells were increased in the GFP− population, but neither was increased in the GFP+ population, suggesting that Mesp1 induction of hematopoiesis is cell autonomous. Ery-P, primitive erythroid; Ery-D, definitive erythroid; GM, granulocyte-macrophage; GEMM, granulocyte-erythrocyte-megakaryocyte-macrophage; Mac, macrophage Mean ± SEM is shown in panels A and D. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox. See also Figure S2
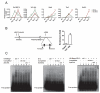
Figure 3. Mesp1 promotes hematopoiesis by regulating Etv2 and Tal1
(A) Quantitative RT-PCR for regulatory hematopoietic and cardiogenic transcription factors during EB differentiation (n=3). Note that EBs treated with the early Mesp1 pulse (green line) rapidly upregulated hematopoietic transcription factors Etv2 and Tal1 at day 3.25 (versus No Dox, gray line), with Tal1 continuing to be elevated by day 4, but expression levels fell back to or below the control (No Dox) level by day 6. On the other hand, the late pulse of Mesp1 (red line) led to a gradual and sustained upregulation of cardiogenic transcription factors Gata4, Tbx5 and Nkx2-5 from day 4 to day 6. (B) Schematic showing the +40k enhancer of Tal1 and the E-box motifs therein (left). The relative locations of a nearby gene Pdzk1ip1 (Map17) and the primer set for PCR detection are also shown. ChIP analysis on day 3 EBs (right, n=3) illustrates that Mesp1 binds to this _Tal1 cis_-regulatory element. Gapdh was used as a control. (C) Electrophoretic mobility shift assay analysis showing that neither Mesp1 (left) nor its co-factor E12 (an isoform of E2A) (middle) alone bound to the E-boxes of the Tal1 +40k enhancer (left), but their dimerization complex could (right). Specific binding and supershift signal are indicated by the arrowhead and the star respectively. ctrl, control (cell lysate from empty vector for lane 2 and IgG antibody for lane 6); mt, mutant (E-box motif mutant competitor); wt, wildtype (wildtype competitor) Mean ± SEM is shown in panels A and B. *, p<0.05; **, p<0.01; ***, p<0.001 versus No Dox. See also Figure S3
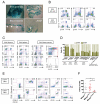
Figure 4. Mesp1 marks progenitors that contribute to both embryonic and adult hematopoiesis
(A) Whole-mount lacZ staining of Mesp1Cre/+;R26fl-stop-lacZ/+ E9.5 embryos and yolk sacs and sections thereof. Note that lacZ staining was observed in cells of mesodermal origin but not cells in the neural crest and the yolk sac endoderm layer. In particular, lacZ+ blood cells (white arrowheads) were found within the heart chambers, the AGM region and the yolk sac blood islands. Bar = 500 μm (upper left) or 100 μm (others). (B) FACS profiles of hematopoietic (Ter119, CD41) and endothelial (Flk-1) markers in Mesp1Cre/+;R26fl-stop-EYFP/+ E9.5 yolk sacs (top row) and embryos (bottom row). (C) FACS profiles of hematopoietic markers in the thymus (far left), the spleen (middle left) and the bone marrow (middle and far right) of adult Mesp1Cre/+;R26REYFP/+ animals. (D) Quantification of EYFP expression by FACS in hematopoietic fractions of various origins in E9.5 and adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals (n=5-7). Note that all hematopoietic lineages examined contain a significant EYFP+ fraction. (E) Mesp1-marked bone marrow cells repopulated and contributed to multiple hematopoietic lineages. EYFP+ (top row) and EYFP− (bottom row) total bone marrow cells from adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals were transplanted to NSG-CD45.1/CD45.1 recipient mice. FACS analysis of the peripheral blood 4 months later showed that almost all blood cells derived from the donor (CD45.2+) but not from the host (CD45.1+) (far left). Note that Mesp1-unlabelled transplanted cells remained EYFP− (bottom row, near left), indicating that Mesp1 is not re-expressed in the adult hematopoietic system. (F) Hematopoietic colony-forming assay showing that yolk sacs of Mesp1Cre/Cre knockout embryos (E8.25-E8.5, equivalent to 6-8 somite pairs) produced fewer hematopoietic colonies than those of Mesp1+/+ and Mesp1Cre/+ embryos. *, p<0.05 AGM, aorta-gonads-mesonephros; BMT, bone marrow transplantation; Lin, lineage cocktail comprising CD3, CD4, CD8, Mac1, B220, Gr-1 and Ter119; HSC, hematopoietic stem cells; NSG-CD45.1/CD45.1, CD45.1/CD45.1 homozygous on the NOD scid gamma background. Mean ± SEM is shown in panel D. See also Figure S4
Figure 5. Mesp1 promotes paraxial mesoderm and myogenic derivatives in the absence of serum-derived factors
(A) Scheme depicting the protocol used to examine the effect of Mesp1 on ES differentiation in serum-free conditions. (B) FACS analysis of mesoderm markers Flk-1 and PDGFRα in day 5 EBs grown under serum-free conditions. (C) Quantitative RT-PCR for various lineage-specific markers in serum-free culture conditions in which Mesp1 was induced from day 3 to day 8 (blue bars) or not induced (gray bars) (n=3). (D) Immunoblot showing upregulation of myogenic proteins upon Mesp1 induction from day 3 to day 8 under serum-free conditions. (E) Immunostaining for myogenic markers, MyoD (left), Myogenin (middle) and MHC (right), in EB-derived cells induced by Mesp1 from day 3-8 cultured in the absence of serum. Areas depicted by the white dotted rectangle are magnified to demonstrate the localization of the nucleus (DAPI) and the nuclear (MyoD and Myogenin) or cytoplasmic (MHC) markers of interest (bottom panels). No MyoD+, Myogenin+ or MHC+ cells were observed in control (No Dox) wells (not shown). Bar = 200 μm (F) Immunostaining showing the presence of multiple nuclei (white arrowheads) in Mesp1-induced ES cell-derived MHC+ myotube. The red channel (MHC) and the blue channel (DAPI) are individually merged with the phase contrast channel to indicate that multiple DAPI+ nuclei are present in the same MHC+ myogenic cell (bottom panels). Bar = 100 μm Mean ± SEM is shown in panel C.
Figure 6. Mesp1-expressing progenitors contribute to the adult satellite cell pool
(A) FACS analysis of TA muscle from Pax7-ZsGreen mice confirming that more than 90% of Lin− α7-integrin+ VCAM-1+ cells are ZsGreen+ (Pax7+). (B) FACS profiles showing the satellite cell marker-gated population (Lin− α7-integrin+ VCAM-1+) and the EYFP content thereof in the facial muscles (far left), the masseter (middle left), the diaphragm (middle right) and the hindlimb muscles (far right) of adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals. (C) Quantification of EYFP expression by FACS in satellite cell marker-gated populations (left: Lin− α7-integrin+ CD34+; right: Lin− α7-integrin+ VCAM-1+) from various muscle groups in adult Mesp1Cre/+;R26fl-stop-EYFP/+ animals (n=4-5). Note that EYFP+ satellite cells are predominant in the facial muscles and masseter, rare in the diaphragm and triceps, and almost undetectable in the back and hindlimb muscles. (D) Immunostaining for MHC in EYFP+ sorted populations gated on satellite cell markers (Lin− α7-integrin+ VCAM-1+) from various muscle groups. Multinucleated MHC+ myotubes co-expressing EYFP+ are observed in all sorted populations, indicating that the gated populations of Mesp1 origin are indeed skeletal myogenic progenitors, and not a non-myogenic subpopulation. Bar = 100 μm. (E) Mesp1-marked satellite cells engrafted and differentiated into muscle fibers. EYFP+ satellite cells (Lin− α7-integrin+ VCAM-1+) isolated from the masseter and TA of Mesp1Cre/+;R26fl-stop-EYFP/+ animals (dystrophin+) were transplanted into the TA of NSG-mdx4Cv recipients (dystrophin−). Immunostaining revealed the presence of dystrophin+ (red) muscle fibers, indicating engraftment, in transplanted (left and middle) but not in control non-transplanted (right) tissues. Bar = 100 μm Lin, lineage cocktail comprising CD31 (endothelial) and CD45 (hematopoietic). Mean ± SEM is shown in panel C. See also Figure S5
Similar articles
- Heterogeneity of Mesp1+ mesoderm revealed by single-cell RNA-seq.
Chan SS, Chan HHW, Kyba M. Chan SS, et al. Biochem Biophys Res Commun. 2016 Jun 3;474(3):469-475. doi: 10.1016/j.bbrc.2016.04.139. Epub 2016 Apr 27. Biochem Biophys Res Commun. 2016. PMID: 27131741 Free PMC article. - Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm.
Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. Kataoka H, et al. Blood. 2011 Dec 22;118(26):6975-86. doi: 10.1182/blood-2011-05-352658. Epub 2011 Sep 12. Blood. 2011. PMID: 21911838 - Mesp1 Marked Cardiac Progenitor Cells Repair Infarcted Mouse Hearts.
Liu Y, Chen L, Diaz AD, Benham A, Xu X, Wijaya CS, Fa'ak F, Luo W, Soibam B, Azares A, Yu W, Lyu Q, Stewart MD, Gunaratne P, Cooney A, McConnell BK, Schwartz RJ. Liu Y, et al. Sci Rep. 2016 Aug 19;6:31457. doi: 10.1038/srep31457. Sci Rep. 2016. PMID: 27538477 Free PMC article. - Developmental relationship between hematopoietic and endothelial cells.
Lugus JJ, Park C, Choi K. Lugus JJ, et al. Immunol Res. 2005;32(1-3):57-74. doi: 10.1385/IR:32:1-3:057. Immunol Res. 2005. PMID: 16106059 Review. - Mesp1: a key regulator of cardiovascular lineage commitment.
Bondue A, Blanpain C. Bondue A, et al. Circ Res. 2010 Dec 10;107(12):1414-27. doi: 10.1161/CIRCRESAHA.110.227058. Circ Res. 2010. PMID: 21148448 Review.
Cited by
- Regulation and evolution of cardiopharyngeal cell identity and behavior: insights from simple chordates.
Kaplan N, Razy-Krajka F, Christiaen L. Kaplan N, et al. Curr Opin Genet Dev. 2015 Jun;32:119-28. doi: 10.1016/j.gde.2015.02.008. Epub 2015 Mar 25. Curr Opin Genet Dev. 2015. PMID: 25819888 Free PMC article. Review. - Cardiac Progenitor Cells from Stem Cells: Learning from Genetics and Biomaterials.
Barreto S, Hamel L, Schiatti T, Yang Y, George V. Barreto S, et al. Cells. 2019 Nov 28;8(12):1536. doi: 10.3390/cells8121536. Cells. 2019. PMID: 31795206 Free PMC article. Review. - Generation of craniofacial myogenic progenitor cells from human induced pluripotent stem cells for skeletal muscle tissue regeneration.
Kim E, Wu F, Wu X, Choo HJ. Kim E, et al. Biomaterials. 2020 Jul;248:119995. doi: 10.1016/j.biomaterials.2020.119995. Epub 2020 Apr 2. Biomaterials. 2020. PMID: 32283390 Free PMC article. - What is a Master Regulator?
Chan SS, Kyba M. Chan SS, et al. J Stem Cell Res Ther. 2013 May 4;3:114. doi: 10.4172/2157-7633.1000e114. J Stem Cell Res Ther. 2013. PMID: 23885309 Free PMC article. No abstract available. - How Mesp1 makes a move.
Kelly RG. Kelly RG. J Cell Biol. 2016 May 23;213(4):411-3. doi: 10.1083/jcb.201604121. Epub 2016 May 16. J Cell Biol. 2016. PMID: 27185831 Free PMC article.
References
- Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am. J. Med. Genet. C Semin. Med. Genet. 2007;145C:109–116. - PubMed
- Agha-Mohammadi S, O’Malley M, Etemad A, Wang Z, Xiao X, Lotze MT. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene Med. 2004;6:817–828. - PubMed
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR055685/AR/NIAMS NIH HHS/United States
- T32 AR007612/AR/NIAMS NIH HHS/United States
- T32 HL069764/HL/NHLBI NIH HHS/United States
- U01 HL100407/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous