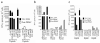Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells - PubMed (original) (raw)
doi: 10.1038/nm.3161. Epub 2013 May 5.
Yong-Chen Lu, Mona El-Gamil, Yong F Li, Colin Gross, Jared Gartner, Jimmy C Lin, Jamie K Teer, Paul Cliften, Eric Tycksen, Yardena Samuels, Steven A Rosenberg
Affiliations
- PMID: 23644516
- PMCID: PMC3757932
- DOI: 10.1038/nm.3161
Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells
Paul F Robbins et al. Nat Med. 2013 Jun.
Abstract
Substantial regressions of metastatic lesions have been observed in up to 70% of patients with melanoma who received adoptively transferred autologous tumor-infiltrating lymphocytes (TILs) in phase 2 clinical trials. In addition, 40% of patients treated in a recent trial experienced complete regressions of all measurable lesions for at least 5 years following TIL treatment. To evaluate the potential association between the ability of TILs to mediate durable regressions and their ability to recognize potent antigens that presumably include mutated gene products, we developed a new screening approach involving mining whole-exome sequence data to identify mutated proteins expressed in patient tumors. We then synthesized and evaluated candidate mutated T cell epitopes that were identified using a major histocompatibility complex-binding algorithm for recognition by TILs. Using this approach, we identified mutated antigens expressed on autologous tumor cells that were recognized by three bulk TIL lines from three individuals with melanoma that were associated with objective tumor regressions following adoptive transfer. This simplified approach for identifying mutated antigens recognized by T cells avoids the need to generate and laboriously screen cDNA libraries from tumors and may represent a generally applicable method for identifying mutated antigens expressed in a variety of tumor types.
Figures
Figure 1. Response of TIL 2098 to candidate epitopes identified from autologous tumors
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 2098 in an overnight co-culture with peptide-pulsed T2 cells that were pulsed individually with the top 61 candidate HLA-A*0201 binding peptides (Supplementary Table 1) identified from 2098 mel, with the exception of peptides 9,10,14,23,24,31,33 and 37, which overlapped with peptides 1,4,4,25,12,15,16 and 4, respectively (NT). Peptides 39-62 (Supplementary Table 1) stimulated the release of 100 pg/ml or less of IFN-γ from TIL 2098 and are not depicted in this graph. The autologous 2098 mel stimulate the release of 10,000 pg/ml of IFN-γ from 2098 TIL in this assay. (b-d) T2 cells were pulsed with tittered doses of the indicated mutated or wild type (WT) peptides, and IFN-γ release measured in an overnight co-culture with TIL 2098. (e) Stable transfected of COS7 cells (COS-A2) expressing HLA-A*0201 were transiently transfected with the indicated transcripts and evaluated for their ability to stimulate IFN-γ release from TIL 2098 in an overnight co-culture. (f) 293 cells were transiently transfected with a construct encoding HLA-*0201 as well as the indicated transcripts and evaluated for their ability to stimulate IFN-γ release from TIL 2098 in an overnight co-culture.
Figure 2. Response of TIL 2369 to candidate epitopes identified from autologous tumors
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 2369 in an overnight co-culture with peptide-pulsed 293-A1 cells that were pulsed individually with the top 56 candidate HLA-A*0101-binding peptides identified from 2369 mel (Supplementary Table 2), with the exception of peptides 35, 39 and 44, which overlapped with peptides 20, 33 and 43, respectively, and were not tested (NT). Peptides 24-56 (Supplementary Table 2) stimulated the release of less than 100 pg/ml of IFN-γ from TIL 2369 and are not depicted in this graph. The autologous 2369 mel stimulated the release of 20,838 pg/ml of IFN-γ from TIL 2369 in this assay. (b,c) 293-A1 cells were pulsed with the indicated concentrations of peptides in an overnight co-culture with TIL 2098, and IFN-γ release measured. d. 293-A1 cells were transiently transfected with the indicated constructs and evaluated for their ability to stimulate IFN-γ release from TIL 2369 in an overnight co-culture.
Figure 3. Response of TIL 3309 to candidate epitopes identified from autologous tumors
(a) A screening assay was carried out to evaluate the release of IFN-γ from TIL 3309 in an overnight co-culture with COS-A11 cells that were pulsed individually with the top 46 candidate HLA-A*1101-binding peptides identified from 3309 mel (Supplementary Table 5). Peptides 37-46 (Supplementary Table 5) stimulated the release of less than 100 pg/ml of IFN-γ from TIL 3309 and are not depicted in this graph. The autologous 3309 mel stimulated the release of 4,600 pg/ml of IFN-γ from TIL 3309 in this assay. (b,c) COS7 cells stably transduced with a retroviral vector expressing HLA-A*1101 (COS-A11) were pulsed with the indicated concentrations of peptides in an overnight co-culture with TIL 3309, and IFN-γ release measured. d. COS-A11 cells were transiently transfected with the indicated constructs and evaluated for their ability to stimulate IFN-γ release from TIL 3309 in an overnight co-culture.
Figure 4. IFN-γ ELISPOT responses of TIL and PBMC obtained prior to and following autologous TIL transfer
Samples of PBMC prior to adoptive TIL transfer as well as samples of PBMC obtained approximately one month following transfer were evaluated along with samples of infused TIL. A pre-treatment sample was not available for subject 1, and the pre-treatment sample analyzed from subject 2 was obtained approximately one month following an adoptive TIL transfer that was administered in the absence of non-myeloablative chemotherapy one month prior to infusion of the TIL analyzed in (b). TIL 2098 from subject 1 was cultured with HLA-A*0201+ COS-7 cells (a), TIL 2369 from subject 2 was cultured with HLA*0101+ COS7 cells (b), and TIL 3309 from subject 3 were cultured with HLA*1101+ COS-7 cells (c) that were pulsed with candidate nonamer peptides for two hours at 370C. T cells were also cultured with autologous as well as allogeneic tumor cells, and in addition, each of the individual T cell populations were stimulated in parallel cultures with PMA plus ionomycin (PMA/I). The numbers of IFN-γ ELISPOTS per 105 T cells generated in responses to PMA/I were as follows: Subject 1 TIL 2098:7,300, subject 1 post transfer PBMC:4,700, Subject 2 TIL 2369:21,800, Subject 2 pre-transfer PBMC:21,800, Subject 2 post-transfer PBMC:11,000, Subject 3 TIL 3309:7,700, Subject 3 pre-transfer PBMC: 12,500, Subject 3 post-transfer PBMC:26,300. TIL 2098 was plated at 2,000 cells per well, TIL 2369 and 3309 were plated at 1,000 cells per well, and PBMC were plated at 100,000 cells per well, whereas all of the groups stimulated with PMA/I were plated at 1,000 cells per well. The error bars represent the mean + and − the standard error of the mean of three replicate wells per group.
Comment in
- Immunotherapy: Adoptive cell therapy simplified.
Kirk R. Kirk R. Nat Rev Clin Oncol. 2013 Jul;10(7):368. doi: 10.1038/nrclinonc.2013.85. Epub 2013 May 21. Nat Rev Clin Oncol. 2013. PMID: 23689751 No abstract available. - Tumor antigenicity revealed.
Savage PA. Savage PA. Trends Immunol. 2014 Feb;35(2):47-8. doi: 10.1016/j.it.2014.01.001. Epub 2014 Jan 15. Trends Immunol. 2014. PMID: 24439426 Free PMC article.
Similar articles
- Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions.
Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, Rosenberg SA, Robbins PF. Lu YC, et al. Clin Cancer Res. 2014 Jul 1;20(13):3401-10. doi: 10.1158/1078-0432.CCR-14-0433. Clin Cancer Res. 2014. PMID: 24987109 Free PMC article. - Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression.
Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, Rosenberg SA. Parkhurst M, et al. Clin Cancer Res. 2017 May 15;23(10):2491-2505. doi: 10.1158/1078-0432.CCR-16-2680. Epub 2016 Nov 8. Clin Cancer Res. 2017. PMID: 27827318 Free PMC article. - Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes.
Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, Li YF, El-Gamil M, Rosenberg SA, Robbins PF. Cohen CJ, et al. J Clin Invest. 2015 Oct 1;125(10):3981-91. doi: 10.1172/JCI82416. Epub 2015 Sep 21. J Clin Invest. 2015. PMID: 26389673 Free PMC article. - Adoptive T-cell therapy using autologous tumor-infiltrating lymphocytes for metastatic melanoma: current status and future outlook.
Wu R, Forget MA, Chacon J, Bernatchez C, Haymaker C, Chen JQ, Hwu P, Radvanyi LG. Wu R, et al. Cancer J. 2012 Mar-Apr;18(2):160-75. doi: 10.1097/PPO.0b013e31824d4465. Cancer J. 2012. PMID: 22453018 Free PMC article. Review. - The use of melanosomal proteins in the immunotherapy of melanoma.
Kawakami Y, Robbins PF, Wang RF, Parkhurst M, Kang X, Rosenberg SA. Kawakami Y, et al. J Immunother. 1998 Jul;21(4):237-46. doi: 10.1097/00002371-199807000-00001. J Immunother. 1998. PMID: 9672845 Review.
Cited by
- Evolving synergistic combinations of targeted immunotherapies to combat cancer.
Melero I, Berman DM, Aznar MA, Korman AJ, Pérez Gracia JL, Haanen J. Melero I, et al. Nat Rev Cancer. 2015 Aug;15(8):457-72. doi: 10.1038/nrc3973. Nat Rev Cancer. 2015. PMID: 26205340 Review. - Targeting the lipid kinase PIKfyve upregulates surface expression of MHC class I to augment cancer immunotherapy.
Bao Y, Qiao Y, Choi JE, Zhang Y, Mannan R, Cheng C, He T, Zheng Y, Yu J, Gondal M, Cruz G, Grove S, Cao X, Su F, Wang R, Chang Y, Kryczek I, Cieslik M, Green MD, Zou W, Chinnaiyan AM. Bao Y, et al. Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2314416120. doi: 10.1073/pnas.2314416120. Epub 2023 Nov 27. Proc Natl Acad Sci U S A. 2023. PMID: 38011559 Free PMC article. - Clinical protocol phase II study of tumor infiltrating lymphocytes in advanced tumors with alterations in the SWI/SNF complex: the TILTS study.
Martin-Liberal J, Garralda E, García-Donas J, Soto-Castillo JJ, Mussetti A, Codony C, Martin-Lluesma S, Muñoz S, Galvao V, Lostes J, Rotxes M, Prat-Vidal C, Palomero J, Muñoz A, Moreno R, García Del Muro X, Sureda A, Alemany R, Gros A, Piulats JM. Martin-Liberal J, et al. Future Oncol. 2024;20(32):2437-2445. doi: 10.1080/14796694.2024.2385287. Epub 2024 Aug 12. Future Oncol. 2024. PMID: 39129675 - Detecting Tumor Antigen-Specific T Cells via Interaction-Dependent Fucosyl-Biotinylation.
Liu Z, Li JP, Chen M, Wu M, Shi Y, Li W, Teijaro JR, Wu P. Liu Z, et al. Cell. 2020 Nov 12;183(4):1117-1133.e19. doi: 10.1016/j.cell.2020.09.048. Epub 2020 Oct 22. Cell. 2020. PMID: 33096019 Free PMC article. - Mass spectrometric analysis of the HLA class I peptidome of melanoma cell lines as a promising tool for the identification of putative tumor-associated HLA epitopes.
Gloger A, Ritz D, Fugmann T, Neri D. Gloger A, et al. Cancer Immunol Immunother. 2016 Nov;65(11):1377-1393. doi: 10.1007/s00262-016-1897-3. Epub 2016 Sep 6. Cancer Immunol Immunother. 2016. PMID: 27600516 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical