Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori - PubMed (original) (raw)
Review
Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori
Nina R Salama et al. Nat Rev Microbiol. 2013 Jun.
Abstract
The bacterial pathogen Helicobacter pylori has co-evolved with humans and colonizes approximately 50% of the human population, but only causes overt gastric disease in a subset of infected hosts. In this Review, we discuss the pathogenesis of H. pylori and the mechanisms it uses to promote persistent colonization of the gastric mucosa, with a focus on recent insights into the role of the virulence factors vacuolating cytotoxin (VacA), cytotoxin-associated gene A (CagA) and CagL. We also describe the immunobiology of H. pylori infection and highlight how this bacterium manipulates the innate and adaptive immune systems of the host to promote its own persistence.
Figures
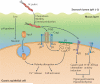
Figure 1. H. pylori colonization and persistence factors
During initial infection of the stomach lumen, urease-dependent ammonia production locally raises the pH, which promotes bacterial survival and solubilizes the mucus gel to facilitate bacterial motility. Chemotaxis(driven by pH and possibly other gradients) and helical rod shape promote flagellar-based motility away from the acidic lumen to H. pylori's preferred niche, on and adjacent to gastric epithelial cells. SabA, BabA and other variably expressed adhesins may shift the balance to cell-associated bacteria. Inset: cell-associated bacteria alter gastric epithelial cell behavior through VacA, CagA and CagL which all have multiple cellular targets. This includes CagA and VacA dependent disruption of cell polarity that can promote iron acquisition or cell extrusion, CagA and CagL dependent induction of chemokines and/or the gastric hormone gastrin, CagL dependent inhibition of acid secretion by the H+/K+ ATPase, and affects on proliferation, apoptosis and differentiation mediated by all three effectors. As noted in the main text, in addition to CagL (depicted), CagA and CagY have also been shown to bind α5β1 integrins although the precise interaction surface remains to be determined. fla, flagella; che, chemotaxis; ure, urease; T4SS, Cag Type IV secretion system; PS, phosphatidylserine; α5β1 and αvβ5, indicated integrin subunits.
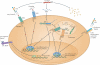
Figure 2. H. pylori subversion of innate immune recognition
H. pylori harbors PAMPs that have evolved to evade detection by pro-inflammatory TLRs. H. pylori expresses tetra-acylated LPS, which is less bioactive than the hexa-acylated form typical of other Gram-negative pathogens due to specific lipid A modifications that prevent detection by TLR4. H. pylori flagella are not detected by TLR5 due to mutations in the TLR5 binding site of flagellin. The bacterium's DNA, as well as an as yet uncharacterized PAMP (and possibly H. pylori LPS) are detected by TLRs 9 and 2, respectively; these TLRS predominantly activate anti-inflammatory signalling pathways and anti-inflammatory IL-10 expression. 5‘ triphosphorylated RNA is detected by the RLR RIG-I, which activates the transcription factors IRF3 and IRF7 to induce type I IFN expression, and is potentially detected also by TLR8 in endosomes. H. pylori's fucosylated DC-SIGN ligands suppress activation of the signalling pathways downstream of this CLR and activate anti-inflammatory genes. Please note that not all depicted TLRs, RLRs and CLRs are necessarily expressed by the same cell type; only one generic cell type is shown here for simplicity. DD, death domain; TIR, Toll/Interleukin-1 receptor domain; CARD, caspase activation and recruitment domain; MyD88, myeloid differentiation primary response gene 88; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3 grabbing non-integrin; SRC, steroid receptor coactivator.
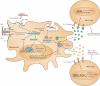
Figure 3. H. pylori activation of NLRs, NF-κB signalling and caspase-1
H. pylori peptidoglycan is delivered to the cytoplasmic NLR Nod1 through either the type IV secretion system (T4SS; via its interaction with α5β1 integrin at cholesterol-rich lipid rafts) or via outer membrane vesicles. Activated Nod1 induces the AP1/NF-κB-dependent expression of pro-inflammatory cytokines and defensins, and the IRF3/7-dependent expression of type I IFNs. Additional unidentified H. pylori NLR ligands activate the inflammasome to induce autoproteolytic pro-caspase-1 cleavage and the subsequent processing and release of mature IL-1β and IL-18. IL-18 binds to its receptor on naive T-cells and promotes FoxP3-dependent Treg differentiation and immune tolerance, which in turn prevents clearance and ensures persistent colonization of H. pylori. In contrast,IL-1β binding to its receptor induces T-bet- and RORγT-dependent Th1 and Th17 differentiation and the expression of the respective signature cytokines IFN-γ and IL-17. Please note that the pictured innate immune cell is a dendritic cell, whereas peptidoglycan-induced Nod1 signalling has been demonstrated in gastric epithelial cells. OMVs, outer membrane vesicles; ASC, apoptosis-associated speck-like protein containing a carboxy-terminal CARD domain; NOD, nucleotide-binding oligomerization domain; LRR, leucine-rich repeat domain; RICK, receptor-interacting serine/threonine kinase; MAPK, MAP kinase.
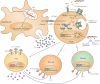
Figure 4. H. pylori impairs T-cell-mediated immunity via direct and indirect mechanisms
All strains of H. pylori express the secreted virulence factors VacA and GGT to directly inhibit T-cell activation, proliferation and effector functions. Hexameric VacA binds to the β2 integrin subunit of the heterodimeric transmembrane receptor LFA-1; the receptor complex is internalized upon protein kinase C-mediated serine/threonine phosphorylation of the β2 integrin cytoplasmic tail. Cytoplasmic VacA prevents nuclear translocation of NF-AT by inhibiting its dephosphorylation by the Ca2+/calmodulin-dependent phosphatase calcineurin, and thereby blocks IL-2 production and subsequent T-cell activation and proliferation. GGT arrests T-cells in the G1 phase of the cell cycle and thus prevents their proliferation. Both VacA and GGT also indirectly prevent T-cell immunity via re-programming of DCs; VacA/GGT-exposed DCs produce IL-10, and induce the FoxP3- and contact-dependent differentiation of T-cells into regulatory T-cells while at the same time preventing Th1 and Th17 differentiation. Depicted interactions at the T-cell/DC synapse include MHCII binding to the T-cell receptor and binding of costimulatory molecules CD80/CD86 to CD28. DC-derived and/or Treg-derived IL-10 further suppresses Th1 and Th17 effector functions. Note that the direct effects of VacA on T-cells appear to be human-specific, whereas indirect effects of VacA and GGT on T-cells via DCs have only been documented in the murine system. LFA-1, lymphocyte function-associated antigen-1; NF-AT- nuclear factor of activated T-cells; GGT, γ-glutamyl-transpeptidase; CnA,B, calcineurin A and B subunits; CaM, calmodulin; RORγT, retinoid-related orphan receptor γT; T-bet, T-box transcription factor.
Similar articles
- Structural modifications of Helicobacter pylori lipopolysaccharide: an idea for how to live in peace.
Chmiela M, Miszczyk E, Rudnicka K. Chmiela M, et al. World J Gastroenterol. 2014 Aug 7;20(29):9882-97. doi: 10.3748/wjg.v20.i29.9882. World J Gastroenterol. 2014. PMID: 25110419 Free PMC article. Review. - Immune evasion strategies used by Helicobacter pylori.
Lina TT, Alzahrani S, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE. Lina TT, et al. World J Gastroenterol. 2014 Sep 28;20(36):12753-66. doi: 10.3748/wjg.v20.i36.12753. World J Gastroenterol. 2014. PMID: 25278676 Free PMC article. Review. - Mechanisms of persistence, innate immune activation and immunomodulation by the gastric pathogen Helicobacter pylori.
Zhang X, Arnold IC, Müller A. Zhang X, et al. Curr Opin Microbiol. 2020 Apr;54:1-10. doi: 10.1016/j.mib.2020.01.003. Epub 2020 Jan 31. Curr Opin Microbiol. 2020. PMID: 32007716 Review. - The Human Stomach in Health and Disease: Infection Strategies by Helicobacter pylori.
Robinson K, Letley DP, Kaneko K. Robinson K, et al. Curr Top Microbiol Immunol. 2017;400:1-26. doi: 10.1007/978-3-319-50520-6_1. Curr Top Microbiol Immunol. 2017. PMID: 28124147 Review. - Immune Evasion Strategies and Persistence of Helicobacter pylori.
Mejías-Luque R, Gerhard M. Mejías-Luque R, et al. Curr Top Microbiol Immunol. 2017;400:53-71. doi: 10.1007/978-3-319-50520-6_3. Curr Top Microbiol Immunol. 2017. PMID: 28124149 Review.
Cited by
- Spatial and Temporal Shifts in Bacterial Biogeography and Gland Occupation during the Development of a Chronic Infection.
Keilberg D, Zavros Y, Shepherd B, Salama NR, Ottemann KM. Keilberg D, et al. mBio. 2016 Oct 11;7(5):e01705-16. doi: 10.1128/mBio.01705-16. mBio. 2016. PMID: 27729513 Free PMC article. - Cytoskeletal components can turn wall-less spherical bacteria into kinking helices.
Lartigue C, Lambert B, Rideau F, Dahan Y, Decossas M, Hillion M, Douliez JP, Hardouin J, Lambert O, Blanchard A, Béven L. Lartigue C, et al. Nat Commun. 2022 Nov 14;13(1):6930. doi: 10.1038/s41467-022-34478-0. Nat Commun. 2022. PMID: 36376306 Free PMC article. - Helicobacter pylori chronic infection and mucosal inflammation switches the human gastric glycosylation pathways.
Magalhães A, Marcos-Pinto R, Nairn AV, Dela Rosa M, Ferreira RM, Junqueira-Neto S, Freitas D, Gomes J, Oliveira P, Santos MR, Marcos NT, Xiaogang W, Figueiredo C, Oliveira C, Dinis-Ribeiro M, Carneiro F, Moremen KW, David L, Reis CA. Magalhães A, et al. Biochim Biophys Acta. 2015 Sep;1852(9):1928-39. doi: 10.1016/j.bbadis.2015.07.001. Epub 2015 Jul 2. Biochim Biophys Acta. 2015. PMID: 26144047 Free PMC article. - Current and Future Perspectives in the Diagnosis and Management of Helicobacter pylori Infection.
Shatila M, Thomas AS. Shatila M, et al. J Clin Med. 2022 Aug 30;11(17):5086. doi: 10.3390/jcm11175086. J Clin Med. 2022. PMID: 36079015 Free PMC article. Review. - Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium.
Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA, Kuo CJ, Guillemin K, Amieva MR. Huang JY, et al. Cell Host Microbe. 2015 Aug 12;18(2):147-56. doi: 10.1016/j.chom.2015.07.002. Cell Host Microbe. 2015. PMID: 26269952 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical