Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells - PubMed (original) (raw)
Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells
Kai Fu et al. Nat Commun. 2013.
Abstract
CD25, the alpha chain of the interleukin-2 receptor, is expressed in activated T cells and has a significant role in autoimmune disease and tumorigenesis; however, the mechanisms regulating transcription of CD25 remain elusive. Here we identify the Src-associated substrate during mitosis of 68 kDa (Sam68) as a novel non-Rel component in the nuclear factor-kappaB (NF-κB) complex that confers CD25 transcription. Our results demonstrate that Sam68 has an essential role in the induction and maintenance of CD25 in T cells. T-cell receptor engagement triggers translocation of the inhibitor of NF-κB kinase alpha (IKKα) from the cytoplasm to the nucleus, where it phosphorylates Sam68, causing complex formation with NF-κB in the nucleus. These findings reveal the important roles of KH domain-containing components and their spatial interactions with IKKs in determining the binding targets of NF-κB complexes, thus shedding novel insights into the regulatory specificity of NF-κB.
Conflict of interest statement
Competing financial interests
The authors declare that they have no conflict of interest.
Figures
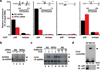
Figure 1. Recognition of CD25 κB DNA requires Rel components but does not depend on RPS3
(a) Jurkat cells transfected with nonspecific (NS) or RPS3 siRNAs were stimulated with αCD3/CD28 (1 µg/ml each). The recruitment of endogenous p65 to the promoters of NFKBIAIL8ACTB, and CD25 genes were analyzed by chromatin immunoprecipitation (ChIP) with either isotype or p65 antibodies. Diagrams inside show the sequences and locations of κB sites in corresponding promoters. The p65-bound DNA was analyzed by quantitative real-time PCR (primers, above diagramed) and normalized to input DNA (mean ± S.D., n = 3). (b–c) Jurkat cells were left untreated (−) or stimulated (+) as in (a), after transfection with siRNAs. The nuclear extracts were analyzed by EMSA with a 32P-labeled NFKBIA κB probe (b) or CD25 κB probe (c). The nonspecific bands and corresponding κB DNA complexes were labeled with asterisks and arrowheads, respectively. In (c), the supershift CD25 κB bands by isotype (iso), p65 (65), or p50 (50) antibodies were labeled with an open arrowhead. (d) Recombinant p65 and p50 proteins (RP) or nuclear extracts (NE) from HUT102 cells was subjected to an EMSA with a 32P-labeled CD25 κB double-stranded DNA, in some cases with 100-fold unlabeled CD25 κB oligonucleotide competitor (CC). The CD25 κB site specific NF-κB DNA binding complex is labeled with a triangle. The amount of p65 and p50 proteins in recombinant proteins and nuclear extracts were immunoblotted (IB) with indicated antibodies.
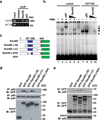
Figure 2. Sam68 is a novel p65-interacting protein in the NF-κB complex binding to the CD25 κB site
(a) The cell surface CD25 expression in resting Jurkat A3 cells and HUT102 cells was stained with isotype antibody or PE-conjugated CD25 antibody, and analyzed by FACS. (b) Nuclear extracts from HUT102 cells were immunoblotted (IB) directly or after immunoprecipitation (IP) with isotype (iso) or p65 antibody for Sam68. (c) The purification scheme for proteins specifically recognizing CD25 κB DNA by gel filtration and DNA affinity purification in the HUT102 cells. (d) Nuclear extracts prepared from HUT102 cells were separated on a Superose 6 gel filtration column. The fractions 14 to 36 were interrogated for CD25 κB DNA binding activity in EMSAs. The unfractionated nuclear extract (NE) was also analyzed with 32P-labeled CD25 κB probe or with 100-fold unlabeled CD25 κB oligonucleotide competitors (CC). The CD25 κB complex was indicated by a triangle and the elution positions of protein standards are indicated. (e) The Superose 6 fractions 14 through 36 were immunoblotted (IB) with p65, p50, and Sam68 antibodies and the elution positions of protein standards are indicated. NE indicates the unfractionated nuclear extract. (f) Schematic diagram of vector double-stranded DNA or that contains 5 copies of CD25 κB sites. Immunoblot (IB) of the elution after DNA affinity purification for indicated proteins from pooled fractions from 24 to 30 harboring high CD25 κB DNA binding activity.
Figure 3. Sam68 is an integral component in the NF-κB complex binding to the CD25 κB site
(a) HUT102 cells were used for ChIP assays with indicated antibodies. The recruitment of indicated proteins to the κB region of CD25 promoter or ACTB promoter in the immunoprecipitates was detected by PCR. (b) Nuclear extracts of HUT102 cells (lanes 7–10) and Jurkat cells (lanes 1–6) treated with (+) or without (−) PMA (50 ng/ml, 30 min) were analyzed by EMSA with 32P-labeled CD25 κB oligonucleotides. Supershift analysis was conducted with p65 or different doses of Sam68 antibodies (lanes 3–5 and 8–10). CD25 κB DNA binding complexes in Jurkat cells and HUT102 cells are labeled with filled triangles, and the supershifted bands and nonspecific bands are labeled with an open triangle and an asterisk, respectively. (c) Schematic diagram of the full-length and truncated mutants (ΔN lacks residues 1–102, ΔC lacks residues 347–443, and ΔKH lacks residues 165–224) of Sam68 fused with EGFP. The hnRNP K homology (KH) domain (residues 157–256) and nuclear localization signal (NLS) are labeled in blue and red, respectively. (d–e) Whole cell lysates from HEK293T cells transfected with GFP vehicle or indicated GFP-fused full-length or truncated Sam68, were immunoblotted (IB) directly or after immunoprecipitation (IP) with p65 antibody (d) or GFP antibody (e) for indicated proteins.
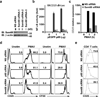
Figure 4. The critical role of Sam68 in the expression of the CD25 gene in T cells
(a) Jurkat cells were transfected with nonspecific (NS) or three different Sam68 specific siRNAs. 72 hr later, the knockdown of Sam68 was examined by immunoblot (IB) for Sam68 with β-actin as a loading control. (b) NF-κB luciferase assay (mean ± S.D., n = 3) of Jurkat cells transfected with nonspecific (NS) or Sam68 siRNAs, together with indicated amount of p65 plasmids and a luciferase reporter gene driven by 5 × CD25 κB sites. (c) Real-time PCR quantization (mean ± S.D., n = 3) of mRNA level of CD25 normalized to GAPDH in Jurkat cells silenced with NS or pooled Sam68 siRNAs and stimulated by 50 ng/ml PMA plus 1.5 µM ionomycin (PMA/I) for indicated time. (d) Human peripheral blood T lymphocytes were transfected with nonspecific (NS), p65, or Sam68 siRNAs and then stimulated with PMA/I or left unstimulated (Unstim). Flow cytometry histograms of T cells with CD25 induction and carboxyfluorescein succinimidyl ester (CFSE) dilution were assessed 12 hr and 72 hr later, respectively. The percentage of CD25+ cells and of dividing cells is shown. (e) Flow-sorted in vitro activated human CD4+CD25+ cells were transfected with nonspecific (NS), p65, or Sam68 siRNAs. The surface CD25 expression was analyzed at 72 hr, and the percentage of CD25+ cells is shown.
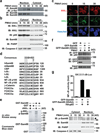
Figure 5. Knockdown of Sam68 does not impair the TCR-engagement-induced NF-κB signaling and p65 nuclear translocation
(a) Jurkat cells expressing either nonspecific (NS) or Sam68 specific siRNA were stimulated with anti-CD3/CD28 (1 µg/ml each, top panel) or 50 ng/ml PMA plus 1.5 µM ionomycin (PMA/I, bottom panel) for indicated periods. Whole cell lysates were extracted and immunoblotted (IB) for IκBα and Sam68, with β-actin as a loading control. (b) Immunoblots using indicated antibodies of cytosolic and nuclear fractions derived from Jurkat cells transfected with indicated siRNAs and left unstimulated or stimulated with PMA/I for indicated periods. PKCθ and PARP served as cytosolic and nuclear markers, respectively. (c) Confocal micrographs of Jurkat cells silenced and stimulated as in (b). The fixed cells were stained for Sam68, p65, and nuclei. (d) Percentage (mean ± S.D., n = 3) of Jurkat cells (n > 200) with nuclear p65 with nonspecific or Sam68 specific siRNA expression was quantified.
Figure 6. IKKα phosphorylates Sam68 in the nucleus during TCR engagement-induced NF-κB activation
(a) Jurkat cells stimulated with PMA plus ionomycin (PMA/I) for indicated periods were fractionated into cytosolic and nuclear subcellular fractions, and then analyzed by immunoprecipitation (IP) and immunoblotted (IB) with antibodies specific for Sam68 and serine-phosphorylated proteins (p-Ser). (b) Cytosolic and nuclear fractions derived from Jurkat cells stimulated as indicated were directly immunoblotted (IB) for indicated proteins. Full-length Caspase 3 and PARP served as cytosolic and nuclear markers, respectively. (c) Confocal micrographs of Jurkat cells stimulated as indicated, and the fixed cells were stained for IKKα, IKKβ, and nuclei. (d) Consensus motif of IKK phosphorylation sites. The sequence of Sam68 is aligned with those of CBP, IκBs, and other known substrates for comparison. The conserved phosphorylation motif for IKK is highlighted in gray (Ψ, hydrophobic amino acid; X, any amino acid). The numbers at right show the position in the amino acid sequence of the last residues depicted. (e) Autoradiograph (upper panel) and Coomassie blue-stained gel (bottom panel) of in vitro kinase assays performed with recombinant GST or GST-Sam68 protein using recombinant human IKKα as kinase. The phosphorylated IKKα and Sam68 were labeled respectively. (f) Whole cell lysates from Jurkat cells expressing either GFP vehicle or indicated GFP-fused wild type or mutant Sam68, were stimulated with PMA/I for 30 min and immunoblotted (IB) directly or after immunoprecipitation (IP) with GFP antibody for indicated proteins. (g) NF-κB luciferase assay (mean ± S.D., n = 3) of Jurkat cells transfected with p65 and indicated GFP-fused wild type or mutant Sam68 plasmids together with a luciferase reporter gene driven by 5 × CD25 κB sites. (h) Cytosolic and nuclear fractions derived from Jurkat cells stimulated as indicated were directly immunoblotted (IB) for Sam68 phosphorylation with antibodies specific for S113/S117 phosphorylated Sam68 and general Sam68. Full-length Caspase 3 and PARP served as cytosolic and nuclear markers, respectively. Phosphorylated Sam68 is labeled by a triangle, and asterisks label nonspecific bands.
Similar articles
- Forkhead transcription factor FOXP3 upregulates CD25 expression through cooperation with RelA/NF-κB.
Camperio C, Caristi S, Fanelli G, Soligo M, Del Porto P, Piccolella E. Camperio C, et al. PLoS One. 2012;7(10):e48303. doi: 10.1371/journal.pone.0048303. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144749 Free PMC article. - Participation of the E3-ligase TRIM13 in NF-κB p65 activation and NFAT-dependent activation of c-Rel upon T-cell receptor engagement.
Hatchi EM, Poalas K, Cordeiro N, N'Debi M, Gavard J, Bidère N. Hatchi EM, et al. Int J Biochem Cell Biol. 2014 Sep;54:217-22. doi: 10.1016/j.biocel.2014.07.012. Epub 2014 Aug 1. Int J Biochem Cell Biol. 2014. PMID: 25088585 - Sam68 modulates apoptosis of intestinal epithelial cells via mediating NF-κB activation in ulcerative colitis.
Qian J, Zhao W, Miao X, Li L, Zhang D. Qian J, et al. Mol Immunol. 2016 Jul;75:48-59. doi: 10.1016/j.molimm.2016.05.011. Epub 2016 May 26. Mol Immunol. 2016. PMID: 27235792 - Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity.
Karin M, Ben-Neriah Y. Karin M, et al. Annu Rev Immunol. 2000;18:621-63. doi: 10.1146/annurev.immunol.18.1.621. Annu Rev Immunol. 2000. PMID: 10837071 Review. - Regulation and function of IKK and IKK-related kinases.
Häcker H, Karin M. Häcker H, et al. Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review.
Cited by
- Sam68/KHDRBS1 is critical for colon tumorigenesis by regulating genotoxic stress-induced NF-κB activation.
Fu K, Sun X, Wier EM, Hodgson A, Liu Y, Sears CL, Wan F. Fu K, et al. Elife. 2016 Jul 25;5:e15018. doi: 10.7554/eLife.15018. Elife. 2016. PMID: 27458801 Free PMC article. - NF-κB Signaling Pathways in Osteoarthritic Cartilage Destruction.
Choi MC, Jo J, Park J, Kang HK, Park Y. Choi MC, et al. Cells. 2019 Jul 17;8(7):734. doi: 10.3390/cells8070734. Cells. 2019. PMID: 31319599 Free PMC article. Review. - Tumor suppressor NME1/NM23-H1 modulates DNA binding of NF-κB RelA.
Shahabi S, Maurya M, Subramaniam S, Ghosh G. Shahabi S, et al. Res Sq [Preprint]. 2024 Oct 14:rs.3.rs-5242004. doi: 10.21203/rs.3.rs-5242004/v1. Res Sq. 2024. PMID: 39483891 Free PMC article. Preprint. - Intrapulmonary administration of purified NEIL2 abrogates NF-κB-mediated inflammation.
Tapryal N, Shahabi S, Chakraborty A, Hosoki K, Wakamiya M, Sarkar G, Sharma G, Cardenas VJ, Boldogh I, Sur S, Ghosh G, Hazra TK. Tapryal N, et al. J Biol Chem. 2021 Jan-Jun;296:100723. doi: 10.1016/j.jbc.2021.100723. Epub 2021 Apr 28. J Biol Chem. 2021. PMID: 33932404 Free PMC article. - Interference with nuclear factor kappaB signaling pathway by pathogen-encoded proteases: global and selective inhibition.
Hodgson A, Wan F. Hodgson A, et al. Mol Microbiol. 2016 Feb;99(3):439-52. doi: 10.1111/mmi.13245. Epub 2015 Nov 5. Mol Microbiol. 2016. PMID: 26449378 Free PMC article. Review.
References
- Kuhn DJ, Dou QP. The role of interleukin-2 receptor alpha in cancer. Front. Biosci. 2005;10:1462–1474. - PubMed
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009110/CA/NCI NIH HHS/United States
- R00 CA137171/CA/NCI NIH HHS/United States
- R01 HL113304/HL/NHLBI NIH HHS/United States
- T32CA009110/CA/NCI NIH HHS/United States
- EP-D-13-052/EPA/United States
- R00CA137171/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous