Simultaneous identification of multiple driver pathways in cancer - PubMed (original) (raw)
Simultaneous identification of multiple driver pathways in cancer
Mark D M Leiserson et al. PLoS Comput Biol. 2013.
Abstract
Distinguishing the somatic mutations responsible for cancer (driver mutations) from random, passenger mutations is a key challenge in cancer genomics. Driver mutations generally target cellular signaling and regulatory pathways consisting of multiple genes. This heterogeneity complicates the identification of driver mutations by their recurrence across samples, as different combinations of mutations in driver pathways are observed in different samples. We introduce the Multi-Dendrix algorithm for the simultaneous identification of multiple driver pathways de novo in somatic mutation data from a cohort of cancer samples. The algorithm relies on two combinatorial properties of mutations in a driver pathway: high coverage and mutual exclusivity. We derive an integer linear program that finds set of mutations exhibiting these properties. We apply Multi-Dendrix to somatic mutations from glioblastoma, breast cancer, and lung cancer samples. Multi-Dendrix identifies sets of mutations in genes that overlap with known pathways - including Rb, p53, PI(3)K, and cell cycle pathways - and also novel sets of mutually exclusive mutations, including mutations in several transcription factors or other genes involved in transcriptional regulation. These sets are discovered directly from mutation data with no prior knowledge of pathways or gene interactions. We show that Multi-Dendrix outperforms other algorithms for identifying combinations of mutations and is also orders of magnitude faster on genome-scale data. Software available at: http://compbio.cs.brown.edu/software.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
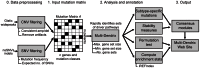
Figure 1. The Multi-Dendrix pipeline.
Multi-Dendrix analyzes integrated mutation data from a variety of sources including single-nucleotide mutations and copy number aberrations. Multiple gene set are identified using a combinatorial optimization approaches. The output is analyzed for subtype-specific mutations and summarized across multiple values of the parameters:  , number of gene sets, and
, number of gene sets, and  , maximum size per gene set.
, maximum size per gene set.
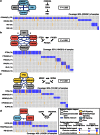
Figure 2. Multi-Dendrix results on the GBM dataset.
(Left) Nodes represent genes in four modules found by Multi-Dendrix using  gene sets of minimize size
gene sets of minimize size  and maximum size
and maximum size  . Genes with “(A)” appended are amplification events, genes with “(D)” appended are deletion events, and genes with no annotation are SNVs. Edges connect genes that appear in the same gene set for more than one value of the parameters, with labels indicating the fraction of parameter values for which the pair of genes appear in the same gene set. Color of nodes indicates membership in three signaling pathways noted in as important for GBM: RB, p53, and RTK/RAS/PI(3)K signaling. Shape of nodes indicates genes whose mutations are associated with specific GBM subtypes, and dashed edges connect genes associated with different subtypes. The direct interactions statistic
. Genes with “(A)” appended are amplification events, genes with “(D)” appended are deletion events, and genes with no annotation are SNVs. Edges connect genes that appear in the same gene set for more than one value of the parameters, with labels indicating the fraction of parameter values for which the pair of genes appear in the same gene set. Color of nodes indicates membership in three signaling pathways noted in as important for GBM: RB, p53, and RTK/RAS/PI(3)K signaling. Shape of nodes indicates genes whose mutations are associated with specific GBM subtypes, and dashed edges connect genes associated with different subtypes. The direct interactions statistic  of this collection of gene sets is significant (
of this collection of gene sets is significant ( ). (Middle) Known interactions between proteins in each set and
). (Middle) Known interactions between proteins in each set and  -value for observed number of interactions. (Right) Mutation matrix for each of four modules with mutual exclusive (blue) and co-occurring mutations (orange).
-value for observed number of interactions. (Right) Mutation matrix for each of four modules with mutual exclusive (blue) and co-occurring mutations (orange).
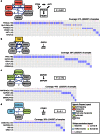
Figure 3. Multi-Dendrix results on the BRCA dataset.
Graphical elements are as described in Figure 2 caption, except for the following. Color of nodes indicates membership in four signaling pathways noted in as important for BRCA: p53 signaling, PI(3)K/AKT signaling, cell cycle checkpoints, and p38-JNK1. The top row of each mutation matrix annotates the subtype of each patient. The regulatory interaction between GATA3 and CDH1 is shown as a dashed line. The direct interactions statistic  of this collection of gene sets is significant (
of this collection of gene sets is significant ( ).
).
Similar articles
- De novo discovery of mutated driver pathways in cancer.
Vandin F, Upfal E, Raphael BJ. Vandin F, et al. Genome Res. 2012 Feb;22(2):375-85. doi: 10.1101/gr.120477.111. Epub 2011 Jun 7. Genome Res. 2012. PMID: 21653252 Free PMC article. - Identifying overlapping mutated driver pathways by constructing gene networks in cancer.
Wu H, Gao L, Li F, Song F, Yang X, Kasabov N. Wu H, et al. BMC Bioinformatics. 2015;16 Suppl 5(Suppl 5):S3. doi: 10.1186/1471-2105-16-S5-S3. Epub 2015 Mar 18. BMC Bioinformatics. 2015. PMID: 25859819 Free PMC article. - Identification of driver pathways in cancer based on combinatorial patterns of somatic gene mutations.
Li HT, Zhang J, Xia J, Zheng CH. Li HT, et al. Neoplasma. 2016;63(1):57-63. doi: 10.4149/neo_2016_007. Neoplasma. 2016. PMID: 26639234 - Computational approaches for the identification of cancer genes and pathways.
Dimitrakopoulos CM, Beerenwinkel N. Dimitrakopoulos CM, et al. Wiley Interdiscip Rev Syst Biol Med. 2017 Jan;9(1):e1364. doi: 10.1002/wsbm.1364. Epub 2016 Nov 11. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 27863091 Free PMC article. Review. - Identifying Epistasis in Cancer Genomes: A Delicate Affair.
van de Haar J, Canisius S, Yu MK, Voest EE, Wessels LFA, Ideker T. van de Haar J, et al. Cell. 2019 May 30;177(6):1375-1383. doi: 10.1016/j.cell.2019.05.005. Cell. 2019. PMID: 31150618 Free PMC article. Review.
Cited by
- Identifying restrictions in the order of accumulation of mutations during tumor progression: effects of passengers, evolutionary models, and sampling.
Diaz-Uriarte R. Diaz-Uriarte R. BMC Bioinformatics. 2015 Feb 12;16:41. doi: 10.1186/s12859-015-0466-7. BMC Bioinformatics. 2015. PMID: 25879190 Free PMC article. - Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations.
Babur Ö, Gönen M, Aksoy BA, Schultz N, Ciriello G, Sander C, Demir E. Babur Ö, et al. Genome Biol. 2015 Feb 26;16(1):45. doi: 10.1186/s13059-015-0612-6. Genome Biol. 2015. PMID: 25887147 Free PMC article. - Analysis, identification and visualization of subgroups in genomics.
Völkel G, Laban S, Fürstberger A, Kühlwein SD, Ikonomi N, Hoffman TK, Brunner C, Neuberg DS, Gaidzik V, Döhner H, Kraus JM, Kestler HA. Völkel G, et al. Brief Bioinform. 2021 May 20;22(3):bbaa217. doi: 10.1093/bib/bbaa217. Brief Bioinform. 2021. PMID: 32954413 Free PMC article. Review. - Adaptively Weighted and Robust Mathematical Programming for the Discovery of Driver Gene Sets in Cancers.
Xu X, Qin P, Gu H, Wang J, Wang Y. Xu X, et al. Sci Rep. 2019 Apr 11;9(1):5959. doi: 10.1038/s41598-019-42500-7. Sci Rep. 2019. PMID: 30976053 Free PMC article. - Review: Precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers.
Nussinov R, Jang H, Tsai CJ, Cheng F. Nussinov R, et al. PLoS Comput Biol. 2019 Mar 28;15(3):e1006658. doi: 10.1371/journal.pcbi.1006658. eCollection 2019 Mar. PLoS Comput Biol. 2019. PMID: 30921324 Free PMC article. Review.
References
- Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols 4: 1073–81. - PubMed
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. - PubMed
Publication types
MeSH terms
Grants and funding
This work is supported by NSF grant IIS-1016648. BJR is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, an Alfred P. Sloan Research Fellowship, and an NSF CAREER Award (CCF-1053753). RS was supported by a research grant from the Israel Science Foundation (grant no. 241/11). MDML was supported by NSF GRFP DGE 0228243. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous