bioNerDS: exploring bioinformatics' database and software use through literature mining - PubMed (original) (raw)
bioNerDS: exploring bioinformatics' database and software use through literature mining
Geraint Duck et al. BMC Bioinformatics. 2013.
Abstract
Background: Biology-focused databases and software define bioinformatics and their use is central to computational biology. In such a complex and dynamic field, it is of interest to understand what resources are available, which are used, how much they are used, and for what they are used. While scholarly literature surveys can provide some insights, large-scale computer-based approaches to identify mentions of bioinformatics databases and software from primary literature would automate systematic cataloguing, facilitate the monitoring of usage, and provide the foundations for the recovery of computational methods for analysing biological data, with the long-term aim of identifying best/common practice in different areas of biology.
Results: We have developed bioNerDS, a named entity recogniser for the recovery of bioinformatics databases and software from primary literature. We identify such entities with an F-measure ranging from 63% to 91% at the mention level and 63-78% at the document level, depending on corpus. Not attaining a higher F-measure is mostly due to high ambiguity in resource naming, which is compounded by the on-going introduction of new resources. To demonstrate the software, we applied bioNerDS to full-text articles from BMC Bioinformatics and Genome Biology. General mention patterns reflect the remit of these journals, highlighting BMC Bioinformatics's emphasis on new tools and Genome Biology's greater emphasis on data analysis. The data also illustrates some shifts in resource usage: for example, the past decade has seen R and the Gene Ontology join BLAST and GenBank as the main components in bioinformatics processing.
Abstract: Conclusions We demonstrate the feasibility of automatically identifying resource names on a large-scale from the scientific literature and show that the generated data can be used for exploration of bioinformatics database and software usage. For example, our results help to investigate the rate of change in resource usage and corroborate the suspicion that a vast majority of resources are created, but rarely (if ever) used thereafter. bioNerDS is available at http://bionerds.sourceforge.net/.
Figures
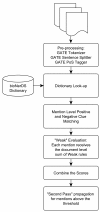
Figure 1
Flowchart of bioNerDS’ name recognition strategy.
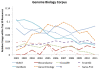
Figure 2
Relative usage of top resources in Genome Biology over time. Highlights the relative usage of some well known bioinformatics resources within the top 50 resources used at document level within Genome Biology.
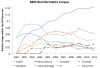
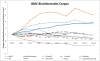
Figure 3
Relative usage of top resources in BMC Bioinformatics over time. Highlights the relative usage of some well known bioinformatics resources within the top 50 resources used at document level within BMC Bioinformatics.
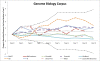
Figure 4
Genome Biology’s upper and lower 95% bounds. Comparison of a resource’s change in relative use, compared to the expected change based on a random walk using a Gaussian distribution fitted to the normalised resource usage changes from a baseline in Year 0 for Genome Biology. The upper and lower 95% bounds are calculated as two standard deviations from the mean.
Figure 5
BMC Bioinformatics’s upper and lower 95% bounds. Comparison of a resource’s change in relative use, compared to the expected change based on a random walk using a Gaussian distribution fitted to the normalised resource usage changes from a baseline in Year 0 for BMC Bioinformatics. The upper and lower 95% bounds are calculated as two standard deviations from the mean.
Figure 6
Genome Biology’s variation in top 50 resource usage. The sum of normalised frequencies against the sum of absolute differences for Genome Biology’s top 50 resource mentions with interesting outliers labelled. The y axis highlights the relative level of use of a resource, whereas the x axis shows the level of variation of tool use across the years 2000 to 2011.
Figure 7
BMC Bioinformatics’s variation in top 50 resource usage. The sum of normalised frequencies against the sum of absolute differences for BMC Bioinformatics’s top 50 resource mentions with interesting outliers labelled. The y axis highlights the relative level of use of a resource, whereas the x axis shows the level of variation of tool use across the years 2000 to 2011.
Similar articles
- Extracting patterns of database and software usage from the bioinformatics literature.
Duck G, Nenadic G, Brass A, Robertson DL, Stevens R. Duck G, et al. Bioinformatics. 2014 Sep 1;30(17):i601-8. doi: 10.1093/bioinformatics/btu471. Bioinformatics. 2014. PMID: 25161253 Free PMC article. - A Survey of Bioinformatics Database and Software Usage through Mining the Literature.
Duck G, Nenadic G, Filannino M, Brass A, Robertson DL, Stevens R. Duck G, et al. PLoS One. 2016 Jun 22;11(6):e0157989. doi: 10.1371/journal.pone.0157989. eCollection 2016. PLoS One. 2016. PMID: 27331905 Free PMC article. - LINNAEUS: a species name identification system for biomedical literature.
Gerner M, Nenadic G, Bergman CM. Gerner M, et al. BMC Bioinformatics. 2010 Feb 11;11:85. doi: 10.1186/1471-2105-11-85. BMC Bioinformatics. 2010. PMID: 20149233 Free PMC article. - Combining literature text mining with microarray data: advances for system biology modeling.
Faro A, Giordano D, Spampinato C. Faro A, et al. Brief Bioinform. 2012 Jan;13(1):61-82. doi: 10.1093/bib/bbr018. Epub 2011 Jun 15. Brief Bioinform. 2012. PMID: 21677032 Review. - Bioinformatics tools and database resources for systems genetics analysis in mice--a short review and an evaluation of future needs.
Durrant C, Swertz MA, Alberts R, Arends D, Möller S, Mott R, Prins P, van der Velde KJ, Jansen RC, Schughart K. Durrant C, et al. Brief Bioinform. 2012 Mar;13(2):135-42. doi: 10.1093/bib/bbr026. Epub 2011 Jul 8. Brief Bioinform. 2012. PMID: 22396485 Free PMC article. Review.
Cited by
- U-Index, a dataset and an impact metric for informatics tools and databases.
Callahan A, Winnenburg R, Shah NH. Callahan A, et al. Sci Data. 2018 Mar 20;5:180043. doi: 10.1038/sdata.2018.43. Sci Data. 2018. PMID: 29557976 Free PMC article. - Resource Disambiguator for the Web: Extracting Biomedical Resources and Their Citations from the Scientific Literature.
Ozyurt IB, Grethe JS, Martone ME, Bandrowski AE. Ozyurt IB, et al. PLoS One. 2016 Jan 5;11(1):e0146300. doi: 10.1371/journal.pone.0146300. eCollection 2016. PLoS One. 2016. PMID: 26730820 Free PMC article. - Recognizing software names in biomedical literature using machine learning.
Wei Q, Zhang Y, Amith M, Lin R, Lapeyrolerie J, Tao C, Xu H. Wei Q, et al. Health Informatics J. 2020 Mar;26(1):21-33. doi: 10.1177/1460458219869490. Epub 2019 Sep 30. Health Informatics J. 2020. PMID: 31566474 Free PMC article. - rAvis: an R-package for downloading information stored in Proyecto AVIS, a citizen science bird project.
Varela S, González-Hernández J, Casabella E, Barrientos R. Varela S, et al. PLoS One. 2014 Mar 13;9(3):e91650. doi: 10.1371/journal.pone.0091650. eCollection 2014. PLoS One. 2014. PMID: 24626233 Free PMC article. - Antibody Exchange: Information extraction of biological antibody donation and a web-portal to find donors and seekers.
Subramanian S, Ganapathiraju MK. Subramanian S, et al. Data (Basel). 2017 Dec;2(4):38. doi: 10.3390/data2040038. Epub 2017 Nov 21. Data (Basel). 2017. PMID: 30498741 Free PMC article.
References
- Cannata N, Merelli E, Altman RB. Time to organize the bioinformatics resourceome. PLoS Comput Biol. 2005;1(7):e76. doi: 10.1371/journal.pcbi.0010076. [ http://www.ncbi.nlm.nih.gov/pubmed/16738704] - DOI - PMC - PubMed
- Wren JD, Bateman A. Databases, data tombs and dust in the wind. Bioinformatics. 2008;24(19):2127–2128. doi: 10.1093/bioinformatics/btn464. [ http://bioinformatics.oxfordjournals.org/cgi/content/abstract/24/19/2127] - DOI - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D J etal. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. - PubMed
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [ http://www.ncbi.nlm.nih.gov/pubmed/17846036] - DOI - PubMed
- Eales JM, Pinney JW, Stevens RD, Robertson DL. Methodology capture discriminating between the “best” and the rest of community practice. BMC Bioinformatics. 2008;9:359. doi: 10.1186/1471-2105-9-359. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2553348. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous