Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia - PubMed (original) (raw)
doi: 10.1007/s00401-013-1147-0. Epub 2013 Jul 2.
Mark Poulter, Tammaryn Lashley, Jonathan D Rohrer, James M Polke, Jon Beck, Natalie Ryan, Davina Hensman, Sarah Mizielinska, Adrian J Waite, Mang-Ching Lai, Tania F Gendron, Leonard Petrucelli, Elizabeth M C Fisher, Tamas Revesz, Jason D Warren, John Collinge, Adrian M Isaacs, Simon Mead
Affiliations
- PMID: 23818065
- PMCID: PMC3753468
- DOI: 10.1007/s00401-013-1147-0
Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia
Pietro Fratta et al. Acta Neuropathol. 2013 Sep.
Abstract
An expanded hexanucleotide repeat in the C9orf72 gene is the most common genetic cause of frontotemporal dementia and amyotrophic lateral sclerosis (c9FTD/ALS). We now report the first description of a homozygous patient and compare it to a series of heterozygous cases. The patient developed early-onset frontotemporal dementia without additional features. Neuropathological analysis showed c9FTD/ALS characteristics, with abundant p62-positive inclusions in the frontal and temporal cortices, hippocampus and cerebellum, as well as less abundant TDP-43-positive inclusions. Overall, the clinical and pathological features were severe, but did not fall outside the usual disease spectrum. Quantification of C9orf72 transcript levels in post-mortem brain demonstrated expression of all known C9orf72 transcript variants, but at a reduced level. The pathogenic mechanisms by which the hexanucleotide repeat expansion causes disease are unclear and both gain- and loss-of-function mechanisms may play a role. Our data support a gain-of-function mechanism as pure homozygous loss of function would be expected to lead to a more severe, or completely different clinical phenotype to the one described here, which falls within the usual range. Our findings have implications for genetic counselling, highlighting the need to use genetic tests that distinguish C9orf72 homozygosity.
Figures
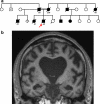
Fig. 1
Family history and clinical features. a Pedigree of the family. Arrow identifies the proband; age of onset is indicated below affected individuals (filled symbols). b Coronal brain MRI T1-weighted scan, showing bilateral widespread atrophy
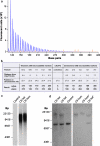
Fig. 2
Presence of a homozygous hexanucleotide expansion in C9orf72. a Results of repeat primed PCR for C9orf72 expansion demonstrating the saw-tooth pattern, typical of the pathological expansion. Repeats are measurable up to 40 hexanucleotide repeats. The size of fluorescently labelled DNA amplicons is shown in base pairs (bp) against the expected asymptotic decay in fluorescence measured in arbitrary units. The GS500 size standard can also be seen with red peaks at 300, 340, 350, 400, 450, 490 and 500 bp. b Results of genetic analysis of the proband that demonstrate homozygosity at all microsatellite marker positions and at rs3849942. Positions of markers are given in megabases relative to chromosome and C9orf72 position. c Southern blot analysis on blood-derived DNA using a probe directed to the hexanucleotide repeat (GGGGCC)5 shows the presence of an expansion in the proband (C9 hom). The expansion is calculated to have a maximum of 3,067, a minimum 830 and mode of 2,297 repeats and is indistinguishable from a C9orf72 heterozygous case (C9 het). d Southern blot analysis on blood and brain (asterisk) derived DNA using a single-copy probe detecting the sequence adjacent to the repeat. The normal allele runs as an 8-kb band and is detectable in controls and C9orf72 expansion heterozygous cases, but is absent in the proband (C9 hom)
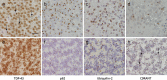
Fig. 3
Immunohistochemical analysis of the neuronal cytoplasmic inclusions in the C9orf72 homozygous case. The ‘_star-like_’ neuronal cytoplasmic inclusions observed in the granule cell layer of the hippocampus (a–d) and the cerebellum (e–h) have been shown to be positive to varying degrees with several antibodies to proteins shown to be associated with C9orf72 cases. TDP-43 immunohistochemistry showed compact neuronal cytoplasmic inclusions (a arrows) together with the normal neuronal nuclear staining pattern. Only the normal nuclear staining pattern was observed in the cerebellum with the TDP-43 antibody (e). p62 immunohistochemisty (b and f) demonstrated large numbers of ‘_star-like_’ inclusions in both the GCL (b) and cerebellum (f). Ubiquilin-2 has been shown to be present in the small ‘_star-like_’ inclusions in heterozygous C9orf72 cases. The homozygous presented here also demonstrated a similar staining pattern with inclusions found in the GCL (c) and cerebellum (g). The newly identified C9RANT protein was also shown to be present in a small proportion of inclusions in the GCL (d) and cerebellum (h arrows). Bar in a represents 20 μm in all panels
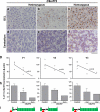
Fig. 4
Histopathological features and C9orf72 expression. p62 immunohistochemistry in the granule cell layer (GCL) of the hippocampus (a–c) and cerebellum (d–f) demonstrating the number of inclusions observed in the heterozygous cases range from mild (a, d) to severe (b, e). The severity of the inclusions observed in the homozygous case (c, f) is shown in both the GCL and cerebellum. Bar represents 100 μm in all panels. Real-time quantitative RT-PCR expression analysis of the three known C9orf72 isoforms—V1, V2, V3 (g, h)—schematically represented in i. Hexanucleotide expansion (red triangle), exons (green), intron (black line). Linear regression analysis (g) between expression and number of normal alleles shows significance for V1 and V2. Reduction of expression in C9orf72 heterozygous cases is significant only for V1 (p = 0.03)
Similar articles
- The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions.
Simón-Sánchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, Smits M, Raaphorst J, van den Berg LH, Schelhaas HJ, De Die-Smulders CE, Majoor-Krakauer D, Rozemuller AJ, Willemsen R, Pijnenburg YA, Heutink P, van Swieten JC. Simón-Sánchez J, et al. Brain. 2012 Mar;135(Pt 3):723-35. doi: 10.1093/brain/awr353. Epub 2012 Feb 1. Brain. 2012. PMID: 22300876 - Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72.
Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB, Rademakers R, Dickson DW. Murray ME, et al. Acta Neuropathol. 2011 Dec;122(6):673-90. doi: 10.1007/s00401-011-0907-y. Epub 2011 Nov 15. Acta Neuropathol. 2011. PMID: 22083254 Free PMC article. - Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p.
Hsiung GY, DeJesus-Hernandez M, Feldman HH, Sengdy P, Bouchard-Kerr P, Dwosh E, Butler R, Leung B, Fok A, Rutherford NJ, Baker M, Rademakers R, Mackenzie IR. Hsiung GY, et al. Brain. 2012 Mar;135(Pt 3):709-22. doi: 10.1093/brain/awr354. Epub 2012 Feb 17. Brain. 2012. PMID: 22344582 Free PMC article. - Prevalence of brain and spinal cord inclusions, including dipeptide repeat proteins, in patients with the C9ORF72 hexanucleotide repeat expansion: a systematic neuropathological review.
Schipper LJ, Raaphorst J, Aronica E, Baas F, de Haan R, de Visser M, Troost D. Schipper LJ, et al. Neuropathol Appl Neurobiol. 2016 Oct;42(6):547-60. doi: 10.1111/nan.12284. Neuropathol Appl Neurobiol. 2016. PMID: 26373655 Review. - Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion.
Babić Leko M, Župunski V, Kirincich J, Smilović D, Hortobágyi T, Hof PR, Šimić G. Babić Leko M, et al. Behav Neurol. 2019 Jan 15;2019:2909168. doi: 10.1155/2019/2909168. eCollection 2019. Behav Neurol. 2019. PMID: 30774737 Free PMC article. Review.
Cited by
- ALS' Perfect Storm: _C9orf72_-Associated Toxic Dipeptide Repeats as Potential Multipotent Disruptors of Protein Homeostasis.
Smeele PH, Cesare G, Vaccari T. Smeele PH, et al. Cells. 2024 Jan 17;13(2):178. doi: 10.3390/cells13020178. Cells. 2024. PMID: 38247869 Free PMC article. Review. - C9orf72-mediated ALS and FTD: multiple pathways to disease.
Balendra R, Isaacs AM. Balendra R, et al. Nat Rev Neurol. 2018 Sep;14(9):544-558. doi: 10.1038/s41582-018-0047-2. Nat Rev Neurol. 2018. PMID: 30120348 Free PMC article. Review. - Microglial crosstalk with astrocytes and immune cells in amyotrophic lateral sclerosis.
Calafatti M, Cocozza G, Limatola C, Garofalo S. Calafatti M, et al. Front Immunol. 2023 Jul 26;14:1223096. doi: 10.3389/fimmu.2023.1223096. eCollection 2023. Front Immunol. 2023. PMID: 37564648 Free PMC article. Review. - Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion.
Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. Waite AJ, et al. Neurobiol Aging. 2014 Jul;35(7):1779.e5-1779.e13. doi: 10.1016/j.neurobiolaging.2014.01.016. Epub 2014 Jan 17. Neurobiol Aging. 2014. PMID: 24559645 Free PMC article. - The Genetics of C9orf72 Expansions.
Gijselinck I, Cruts M, Van Broeckhoven C. Gijselinck I, et al. Cold Spring Harb Perspect Med. 2018 Apr 2;8(4):a026757. doi: 10.1101/cshperspect.a026757. Cold Spring Harb Perspect Med. 2018. PMID: 28130313 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- MR/K000608/1/MRC_/Medical Research Council/United Kingdom
- MC_U123192748/MRC_/Medical Research Council/United Kingdom
- BLAKE/MAR12/815-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G0900421/MRC_/Medical Research Council/United Kingdom
- 091673/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- G0500288/MRC_/Medical Research Council/United Kingdom
- G1000287/MRC_/Medical Research Council/United Kingdom
- MR/K018523/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous