Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens - PubMed (original) (raw)
doi: 10.1038/ni.2679. Epub 2013 Aug 4.
Carlos G Briseño, Jacob S Lee, Dennis Ng, Nicholas A Manieri, Wumesh Kc, Xiaodi Wu, Stephanie R Thomas, Wan-Ling Lee, Mustafa Turkoz, Keely G McDonald, Matthew M Meredith, Christina Song, Cynthia J Guidos, Rodney D Newberry, Wenjun Ouyang, Theresa L Murphy, Thaddeus S Stappenbeck, Jennifer L Gommerman, Michel C Nussenzweig, Marco Colonna, Raphael Kopan, Kenneth M Murphy
Affiliations
- PMID: 23913046
- PMCID: PMC3788683
- DOI: 10.1038/ni.2679
Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens
Ansuman T Satpathy et al. Nat Immunol. 2013 Sep.
Abstract
Defense against attaching-and-effacing bacteria requires the sequential generation of interleukin 23 (IL-23) and IL-22 to induce protective mucosal responses. Although CD4(+) and NKp46(+) innate lymphoid cells (ILCs) are the critical source of IL-22 during infection, the precise source of IL-23 is unclear. We used genetic techniques to deplete mice of specific subsets of classical dendritic cells (cDCs) and analyzed immunity to the attaching-and-effacing pathogen Citrobacter rodentium. We found that the signaling receptor Notch2 controlled the terminal stage of cDC differentiation. Notch2-dependent intestinal CD11b(+) cDCs were an obligate source of IL-23 required for survival after infection with C. rodentium, but CD103(+) cDCs dependent on the transcription factor Batf3 were not. Our results demonstrate a nonredundant function for CD11b(+) cDCs in the response to pathogens in vivo.
Figures
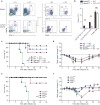
Figure 1. Zbtb46-GFP identifies intestinal cDC populations
(a,b) Lamina propria (a) and MLN (b) cells from _Zbtb46_gfp/+ mice were stained for expression of the indicated markers. Two-color histograms are shown for live cells pre-gated as indicated above the diagram. Numbers represent the percentage of cells within the indicated gate. Data are representative of three independent experiments (n = 9 mice). (c) Small intestine (top) and colon (bottom) sections from _Zbtb46_gfp/+ → wild-type (WT) BM chimeras were analyzed by fluorescence microscopy for expression of B220, CD4, β-catenin, Zbtb46-GFP and DAPI as indicated. Images are representative of two independent experiments (n = 5 mice). Scale bars, 200 μm (Peyer's patch and colonic patch), 100 μm (ILFs and villi). ILF, isolated lymphoid follicle.
Figure 2. Zbtb46+ cDCs are essential for survival after C. rodentium infection
(a, b) _Zbtb46_DTR → WT BM chimeras were treated with 40 ng/g DT at day -3 and day -1 and splenocytes or lamina propria cells were stained for expression of the indicated markers on day 0. (a) Two-color histograms are shown for live cells pre-gated as indicated. (b) Quantification of lamina propria cDC depletion in _Zbtb46_DTR → WT BM chimeras from [a]. Bars represent cDC or macrophage (MΦ) cell numbers per 1 × 106 lamina propria cells. Data are from two independent experiments (error bars, SEM; n = 4 mice, Student's _t_-test). (c,d) The indicated BM chimeras were treated with DT (20 ng/g) one day prior to infection and every third day (5 ng/g) for the remainder of the experiment. Mice were orally inoculated with 2 × 109 c.f.u. C. rodentium and followed for survival (c) and weight loss (d). Data are from two independent experiments (error bars, SEM; n = 8 mice per group, except WT+DT n = 5 mice, survival: log-rank Mantel-Cox test, weight loss: Student's _t_-test). (e,f) Survival (e) and weight loss (f) of mice infected with C. rodentium. Data are from two independent experiments (error bars, SEM; Flt3l+/+ n = 10, _Flt3l_−/− n = 10, Ccr2+/+ n = 5, _Ccr2_−/− n = 4, survival: log-rank Mantel-Cox test, weight loss: Student's _t_-test) * P < 0.05, ** P < 0.001, NS not significant.
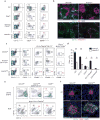
Figure 3. Canonical Notch2 signaling is required for splenic and intestinal CD11b+cDC development
(a) Splenocytes were stained for the indicated markers. Two-color histograms are shown for live cells pre-gated as indicated. Data are representative of three independent experiments (n = 6-8 mice per group). (b) Spleen sections from _Notch2_f/f and _Notch2_vav mice were analyzed by fluorescence microscopy for expression of CD11c, IgD and B220 (top) or CD11c and MadCAM-1 (bottom). Images are representative of two independent experiments (n = 4 mice). Scale bars, 200 μm. (c) Splenocytes were stained for the indicated markers. Data are representative of three independent experiments (n = 7 mice per group). (d) Quantification of cDC development in _Notch2_cKO mice from [c]. Bars represent cDCs per 1 × 106 splenocytes. Data are from three independent experiments (error bars, SEM; n = 7 mice per group, Student's _t_-test). (e) Lamina propria cells were stained for the indicated markers. Data are representative of three independent experiments (n = 3-5 mice per group). (f) Immunofluorescence images of CPs and ILFs from small intestine sections stained for CD11c, CD90 and DAPI. Images are representative of two independent experiments (n = 5 mice). Scale bars, 50 μm. CP, cryptopatch. * P < 0.01, ** P < 0.001.
Figure 4. Notch2 controls terminal differentiation of CD11b+ and DEC205+cDCs
(a) Shown is microarray analysis of sorted cDC subsets from _Notch2_f/f (WT) and _Notch2_cKO (KO) mice. CD11b+ DCs were sorted as MHCII+CD11c+CD24−CD11b+ cells. DEC205+ DCs were sorted as MHCII+CD11c+CD24+CD11b−DEC205+ cells. Colored points represent genes increased more than 2-fold (red) or decreased more than 2-fold (blue) in the indicated cDC subset (left) and then plotted in the complementary subset (right). Data are from two independent experiments (n = 3 biological replicates per cell type in both WT and KO mice, Welch's _t_-test). (b) Left: PCA of WT and KO CD11b+ cDCs and DEC205+ cDCs, analyzed by individual replicate. Proportion of variance: PC1 54.0%, PC2 21.0%. Right: Heat map showing log-transformed expression values from progenitors and cDC subsets derived from the ImmGen database for probesets corresponding to the 20 most positive and negative loadings in PC2. (c) Shown are mean-centered, log-transformed expression values from DC progenitors and subsets for probesets corresponding to the 50 most positive and negative loadings in PC2. Each symbol represents a unique gene (bars, mean; Kruskal-Wallis test, Dunn's multiple comparison test). (d) Expression of selected loadings from [c] were analyzed by gene expression derived from the ImmGen database (left) and by flow cytometry of splenic CD11b+ cDCs in _Ccr2_gfp,_Cx3cr1_gfp, and _Zbtb46_gfp mice (right). Flow cytometry data are representative of three independent experiments (n = 5-6 mice per group). (e) Mixed BM chimeras were generated from CD45.2+_Notch2_vav BM and CD45.1+ WT BM or from CD45.2+_Notch2_f/f and CD45.1+ WT BM. Bone marrow progenitors and splenocytes were analyzed for donor contribution 8-10 weeks following lethal irradiation and transplant. Shown is the contribution of _Notch2_vav BM or _Notch2_f/f BM to each cell-type as a ratio of LSK chimerism in the same animal (% CD45.2+ contribution in cell-type/% CD45.2+ in LSK). Monocyte, neutrophil and cDC chimerism was analyzed in splenocytes. Each symbol represents an individual mouse. Data are from two individual experiments (bars, mean; n = 3-5 mice per group, Student's _t_-test). * P < 0.01, ** P < 0.001.
Figure 5. LTβR signaling mediates the homeostatic expansion of Notch2-dependent cDCs
(a) Splenocytes were stained for the indicated markers. Two-color histograms are shown for live cells pre-gated as indicated. Data are representative of three independent experiments (n = 6 mice per group). (b,c) Mixed BM chimeras were generated with either _Ltbr_−/− and WT BM or with _Ltbr_−/− and _Notch2_cKO BM, and splenocytes or MLN cells were analyzed 8-10 weeks after lethal irradiation and transplant. (b) Shown are two-color histograms for live cells pre-gated as indicated above each diagram (neutrophil: CD11c− CD11b+CD24+). Lower plots indicate representative chimerism ratios (red values). (c) Quantification of mixed chimeras from [b]. Shown is the contribution of _Ltbr_−/− BM to each indicated cell-type as a ratio of neutrophil chimerism in the same animal (% CD45.1+_Ltbr_−/− contribution in cell-type/% CD45.1+_Ltbr_−/− in splenic neutrophils). Each symbol represents an individual mouse. Data are from two independent experiments (bars, mean; n = 2-3 mice per group, Student's _t_-test). (d) Lamina propria cells from the indicated BM chimeras were stained and the contribution of WT or _Ltbr_−/− BM to lamina propria CD103+CD11b+ cDCs was assessed (_Ltbr_−/− = CD45.1+, WT = CD45.1+CD45.2+). (e) Quantification of mixed chimeras generated with _Ltbr_−/− and WT BM from [d] and of chimeras generated with _Ifnar_−/− and WT BM. Chimerism ratio is calculated as (% contribution WT/KO in intestinal cDCs)/(% contribution WT/KO in splenic T cells). Each symbol represents an individual mouse. Data are from five independent experiments (bars, mean ± SEM; _Ltbr_−/− n = 10 mice, _Ifnar_−/− n = 5 mice, Student's _t_-test). * P < 0.05, ** P < 0.01, *** P < 0.001.
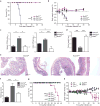
Figure 6. Notch2-dependent CD11b+cDCs are essential for host defense against C. rodentium infection
(a,b) Survival (a) and weight loss (b) of mice orally inoculated with 2 × 109 c.f.u. C. rodentium. Data are from three independent experiments (bars, SEM; n = 9-10 mice per group, Survival: log-rank Mantel-Cox test, weight loss: Student's _t_-test). (c)C. rodentium titers in the spleen and colon 9 days after inoculation. Data are from two independent experiments (bars, SEM; n = 6-7 mice per group, one-way ANOVA, Tukey's multiple comparison test) (d) Colon lengths 9 days after infection. Data are from two independent experiment (bars, SEM; n = 3 mice per group, one-way ANOVA, Tukey's multiple comparison test). (e) H&E staining of colon sections 9 days after infection. Data are representative of two independent experiments (n = 6-7 mice per group). Scale bars, 200 μm. (f) Quantification of histological scores from [e] (bars, SEM; Kruskal-Wallis test, Dunn's multiple comparison test). (g, h) Survival (g) and weight loss (h) of mice infected with C. rodentium. Data are from two independent experiments (bars, SEM; WT n = 8, _Ccr7_−/− n = 5, Irf4−/−n = 6, _Batf2_−/− and Batf2+/+ n = 5, survival: log-rank Mantel-Cox test, weight loss: Student's _t_-test). * P < 0.05, ** P < 0.01, *** P < 0.001, NS not significant.
Figure 7. Notch2-dependent cDCs are dispensable for colonic wound repair
(a) Shown are mean microarray expression values of Ptgs2 for the indicated cell types derived from the Immgen database. Data are assembled from 2-4 replicate arrays (bars, SEM). (b) Whole mount images from colonic wounds 6 days after excision (left). Scale bars, 1 mm. Quantification of the percentage of wound bed sections with incomplete epithelial coverage at day 6 post-excision as measured by whole mount imaging (right). Data are from two independent experiments (bars, mean ± SEM; n = 6 mice per group, Student's _t_-test). Each symbol represents an individual mouse. (c) Shown is the percentage α-SMA loss underlying day 6 wound beds measured in histological sections (gap length/wound bed length). Data are from two independent experiments (bars, SEM; n = 3-6 mice per group). Each symbol represents an individual mouse. (d) Colonic sections from _Notch2_f/f or _Notch2_f/f mice treated with the COX-2 inhibitor NS-398 (5 μg/g) were stained for expression of β-catenin, α-SMA and DAPI on day 2 or day 6 post-excision. Data are representative of two independent experiments (n = 2-3 mice per group). Scale bars, 200 μm. (e) Colonic sections from _Notch2_f/f, _Notch2_cKO or _Batf3_−/− mice were stained for expression of β-catenin, α-SMA, DAPI and F4/80 6 days post-excision. Data are representative of two independent experiments (n = 3-6 mice per group). Scale bars, 200 μm. NS not significant.
Figure 8. Notch2-dependent CD11b+cDCs regulate IL-23-dependent antimicrobial responses to C. rodentium
(a-c) Microarray analysis of colonic cells in mice infected with C. rodentium for 9 days. Shown are M-plots generated using ArrayStar software comparing gene expression between the indicated samples. (b,c) Shown is the average fold change of selected genes from [a]. Selected inflammatory genes increased in expression in _Notch2_cKO (KO) relative to _Notch2_f/f (WT) mice are shown (left). Expression of previously described IL-22-stimulated genes are shown (right). Data are from one independent experiment (n = 2-3 biological replicates per sample). (d) Shown is the normalized expression value of Il22 and Reg3g mRNA determined by qRT-PCR (mRNA/HPRT) for colons isolated from mice infected with C. rodentium for 9 days. Data are from three independent experiments (bars, SEM; n = 6-7 mice per group, one-way ANOVA, Tukey's multiple comparison test). (e) Microarray analysis of colonic cells in mice infected with C. rodentium for 4 days (left). Shown is an M-plot comparing gene expression between the indicated samples. Data are from one independent experiment (n = 2 biological replicates per sample). Shown are representative H&E staining of colons from mice infected for 4 days (right). Data are representative of two independent experiments (n = 4 mice per group). Scale bars, 100 μm. (f) Intracellular IL-22 expression in MLN ILCs from uninfected (UI) or day 4 infected mice. Cells were stimulated ex vivo with PMA and ionomycin, with or without addition of IL-23. Shown are two color histograms for live cells pre-gated as indicated above each diagram. Data are representative of two independent experiments (_n_=4 mice per group). (g) Shown is the normalized expression value of Il23a mRNA determined by qRT-PCR (Il23a/HPRT) from lamina propria CD11b+ cDCs (CD11c+MHCII+CD103+CD11b+) and macrophages (Mac, CD11c+MHCII−CD11b+F4/80+) sorted from uninfected mice or mice infected with C. rodentium for 2 days (C. rod). Data are from two independent experiments (bars, SEM; n = 3 mice per group). (h) Survival of mixed bone marrow chimeras orally inoculated with 2 × 109 C. rodentium 8-10 weeks after lethal irradiation and transplant. Data are from two independent experiments (n = 5-6 per group, log-rank Mantel-Cox test). ND not detected, * P < 0.05, ** P < 0.01, *** P < 0.001, NS not significant.
Comment in
- DCs: a dual bridge to protective immunity.
Sallusto F. Sallusto F. Nat Immunol. 2013 Sep;14(9):890-1. doi: 10.1038/ni.2693. Nat Immunol. 2013. PMID: 23959179 No abstract available.
Similar articles
- IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology.
Aychek T, Mildner A, Yona S, Kim KW, Lampl N, Reich-Zeliger S, Boon L, Yogev N, Waisman A, Cua DJ, Jung S. Aychek T, et al. Nat Commun. 2015 Mar 12;6:6525. doi: 10.1038/ncomms7525. Nat Commun. 2015. PMID: 25761673 Free PMC article. - Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge.
Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu YX. Tumanov AV, et al. Cell Host Microbe. 2011 Jul 21;10(1):44-53. doi: 10.1016/j.chom.2011.06.002. Cell Host Microbe. 2011. PMID: 21767811 Free PMC article. - Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens.
Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Zheng Y, et al. Nat Med. 2008 Mar;14(3):282-9. doi: 10.1038/nm1720. Epub 2008 Feb 10. Nat Med. 2008. PMID: 18264109 - Group 3 innate lymphoid cells mediate host defense against attaching and effacing pathogens.
Jarade A, Di Santo JP, Serafini N. Jarade A, et al. Curr Opin Microbiol. 2021 Oct;63:83-91. doi: 10.1016/j.mib.2021.06.005. Epub 2021 Jul 15. Curr Opin Microbiol. 2021. PMID: 34274597 Review. - Lymphotoxin organizes contributions to host defense and metabolic illness from innate lymphoid cells.
Upadhyay V, Fu YX. Upadhyay V, et al. Cytokine Growth Factor Rev. 2014 Apr;25(2):227-33. doi: 10.1016/j.cytogfr.2013.12.007. Epub 2013 Dec 24. Cytokine Growth Factor Rev. 2014. PMID: 24411493 Free PMC article. Review.
Cited by
- A simplified method for separating renal MPCs using SLAMF9.
Mikulin JA, Bates BL, Wilson TJ. Mikulin JA, et al. Cytometry A. 2021 Dec;99(12):1209-1217. doi: 10.1002/cyto.a.24469. Epub 2021 Jun 14. Cytometry A. 2021. PMID: 34092043 Free PMC article. - Functions of Murine Dendritic Cells.
Durai V, Murphy KM. Durai V, et al. Immunity. 2016 Oct 18;45(4):719-736. doi: 10.1016/j.immuni.2016.10.010. Immunity. 2016. PMID: 27760337 Free PMC article. Review. - Interleukin-22: immunobiology and pathology.
Dudakov JA, Hanash AM, van den Brink MR. Dudakov JA, et al. Annu Rev Immunol. 2015;33:747-85. doi: 10.1146/annurev-immunol-032414-112123. Epub 2015 Feb 11. Annu Rev Immunol. 2015. PMID: 25706098 Free PMC article. Review. - Fate mapping of dendritic cells.
Poltorak MP, Schraml BU. Poltorak MP, et al. Front Immunol. 2015 May 4;6:199. doi: 10.3389/fimmu.2015.00199. eCollection 2015. Front Immunol. 2015. PMID: 25999945 Free PMC article. Review. - ADP-ribosylating adjuvant reveals plasticity in cDC1 cells that drive mucosal Th17 cell development and protection against influenza virus infection.
Arabpour M, Lebrero-Fernandez C, Schön K, Strömberg A, Börjesson V, Lahl K, Ballegeer M, Saelens X, Angeletti D, Agace W, Lycke N. Arabpour M, et al. Mucosal Immunol. 2022 Apr;15(4):745-761. doi: 10.1038/s41385-022-00510-1. Epub 2022 Apr 13. Mucosal Immunol. 2022. PMID: 35418673 Free PMC article.
References
- Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. - PubMed
- Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. - PubMed
- Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. - PubMed
- Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK071619/DK/NIDDK NIH HHS/United States
- U01 AI095542-01/AI/NIAID NIH HHS/United States
- R01 DE021255-01/DE/NIDCR NIH HHS/United States
- T32 HL007317/HL/NHLBI NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- MOP 67157/CAPMC/ CIHR/Canada
- U01 AI095542/AI/NIAID NIH HHS/United States
- R01 GM55479/GM/NIGMS NIH HHS/United States
- R01 DE021255/DE/NIDCR NIH HHS/United States
- R01 DK064798/DK/NIDDK NIH HHS/United States
- AI076427-02/AI/NIAID NIH HHS/United States
- R01 AI076427/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous