Repeat-associated non-ATG (RAN) translation in neurological disease - PubMed (original) (raw)
Review
Repeat-associated non-ATG (RAN) translation in neurological disease
John D Cleary et al. Hum Mol Genet. 2013.
Abstract
Well-established rules of translational initiation have been used as a cornerstone in molecular biology to understand gene expression and to frame fundamental questions on what proteins a cell synthesizes, how proteins work and to predict the consequences of mutations. For a group of neurological diseases caused by the abnormal expansion of short segments of DNA (e.g. CAG•CTG repeats), mutations within or outside of predicted coding and non-coding regions are thought to cause disease by protein gain- or loss-of-function or RNA gain-of-function mechanisms. In contrast to these predictions, the recent discovery of repeat-associated non-ATG (RAN) translation showed expansion mutations can express homopolymeric expansion proteins in all three reading frames without an AUG start codon. This unanticipated, non-canonical type of protein translation is length-and hairpin-dependent, takes place without frameshifting or RNA editing and occurs across a variety of repeat motifs. To date, RAN proteins have been reported in spinocerebellar ataxia type 8 (SCA8), myotonic dystrophy type 1 (DM1), fragile X tremor ataxia syndrome (FXTAS) and C9ORF72 amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD). In this article, we review what is currently known about RAN translation and recent progress toward understanding its contribution to disease.
Figures
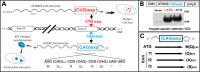
Figure 1.
RAN translation in spinocerebellar ataxia type 8 (SCA8). (A) Prior to the discovery of RAN translation, bidirectional transcription at the SCA8 locus was known to produce RNA foci from the CUG expansion transcript and a polyglutamine expansion protein from the CAG expansion transcript (13,14). The CAG expansion transcript has an unusual short ORF with an ATG initiation codon immediately upstream of the CAG expansion and a series of stop codons immediately after the repeat (14). Both RNA and protein gain-of-function effects are evident in SCA8. (B) To separate the effects of the CUGEXP transcript from the polyGln protein, the ATG immediate upstream of the CAG expanded repeat was mutated in an ATXN8 minigene (11). Unexpectedly, this mutation did not prevent the expression of the polyglutamine protein and was the first indication of RAN translation. (C) Schematic diagram showing CAG-repeat expansion expressing both ATG-initiated polyGln and non-ATG initiated polyGln, polyAla, polySer RAN proteins repeats in all the three reading frames.
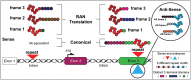
Figure 2.
Model of RAN translation across repeats in coding and non-coding gene regions. Schematic diagram showing mutations located in intronic or exonic regions with expression of distinct RAN proteins in three frames from sense and antisense directions. For expansions in introns, sense and antisense transcripts (not shown) produce RAN proteins with different repeat motifs and distinct C-terminal regions not corresponding to any endogenous proteins. For repeat-expansion mutations located in ORFs, up to six distinct RAN proteins may be produced from sense and antisense transcripts (see upper inset for antisense RAN proteins). The RAN protein expressed in the ORF is predicted to start at or close to the repeat and contain the same C-terminal region as the protein expressed from the canonical ATG-initiated ORF. Variability of RAN proteins will occur when expressed from: sense or antisense transcripts; different repeat motifs and with variations in C-terminal sequences.
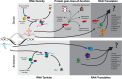
Figure 3.
Potential pathways of pathogenesis of repeat-associated disorders. Bidirectional transcription of an expanded repeat will produced two transcripts (blue = antisense, red = sense), each potentially capable of structure formation and contributions to pathogenesis. In the RNA toxicity model (1st upper and lower panels/light gray), the structures formed by the expanded repeats sequester cellular RNA-binding proteins, thereby interrupting their normal cellular function. The expanded repeats and sequestered proteins may form foci, which may contribute to toxicity or serve a protective function. The proteins sequestered will depend on the structures formed by the RNAs and protein affinity to the structures. For example, expanded CUG transcripts in DM1 sequester the MBNL family of splicing factors which leads to a loss of MBNL function and alternative splicing abnormalities. In the protein gain-/loss-of-function model (second upper panel, medium gray), the ATG-initiated production of expanded proteins may: 1) disrupt or overwhelm cellular pathways (i.e. proteasomes or autophagy) designed to clear aberrant proteins; directly contribute to cellular apoptosis or damage; aggregate or form inclusions that serve a protective function or exacerbate toxicity; or 2) disrupt the normal function of the protein. For example, in huntington's disease, the mutant huntingtin protein disrupts multiple regulatory pathways, including transcription, ubiquitin proteasomal system, autophagy and synaptic transmission. The discovery of RAN translation has added a third potential pathway for disease (upper and lower panels, dark gray). Up to six additional repeat-containing proteins may be produced from the expanded sense and antisense transcripts. RAN proteins may contribute to pathogenesis in a similar or even amplified manner as the protein gain-/loss-of-function pathway. RAN proteins are found within affected patient tissues, suggesting that they contribute to disease.
Similar articles
- Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders.
Cleary JD, Ranum LP. Cleary JD, et al. Curr Opin Genet Dev. 2014 Jun;26:6-15. doi: 10.1016/j.gde.2014.03.002. Epub 2014 May 22. Curr Opin Genet Dev. 2014. PMID: 24852074 Free PMC article. Review. - Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities!
Pearson CE. Pearson CE. PLoS Genet. 2011 Mar;7(3):e1002018. doi: 10.1371/journal.pgen.1002018. Epub 2011 Mar 10. PLoS Genet. 2011. PMID: 21423665 Free PMC article. Review. - Repeat-associated non-ATG (RAN) translation.
Cleary JD, Pattamatta A, Ranum LPW. Cleary JD, et al. J Biol Chem. 2018 Oct 19;293(42):16127-16141. doi: 10.1074/jbc.R118.003237. Epub 2018 Sep 13. J Biol Chem. 2018. PMID: 30213863 Free PMC article. - RAN translation-What makes it run?
Green KM, Linsalata AE, Todd PK. Green KM, et al. Brain Res. 2016 Sep 15;1647:30-42. doi: 10.1016/j.brainres.2016.04.003. Epub 2016 Apr 6. Brain Res. 2016. PMID: 27060770 Free PMC article. Review. - SRSF protein kinase 1 modulates RAN translation and suppresses CGG repeat toxicity.
Malik I, Tseng YJ, Wright SE, Zheng K, Ramaiyer P, Green KM, Todd PK. Malik I, et al. EMBO Mol Med. 2021 Nov 8;13(11):e14163. doi: 10.15252/emmm.202114163. Epub 2021 Sep 20. EMBO Mol Med. 2021. PMID: 34542927 Free PMC article.
Cited by
- RNA-binding proteins in neurodegeneration: Seq and you shall receive.
Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. Nussbacher JK, et al. Trends Neurosci. 2015 Apr;38(4):226-36. doi: 10.1016/j.tins.2015.02.003. Epub 2015 Mar 9. Trends Neurosci. 2015. PMID: 25765321 Free PMC article. Review. - An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I.
López-Martínez A, Soblechero-Martín P, de-la-Puente-Ovejero L, Nogales-Gadea G, Arechavala-Gomeza V. López-Martínez A, et al. Genes (Basel). 2020 Sep 22;11(9):1109. doi: 10.3390/genes11091109. Genes (Basel). 2020. PMID: 32971903 Free PMC article. Review. - Oligonucleotide-based strategies to combat polyglutamine diseases.
Fiszer A, Krzyzosiak WJ. Fiszer A, et al. Nucleic Acids Res. 2014 Jun;42(11):6787-810. doi: 10.1093/nar/gku385. Epub 2014 May 21. Nucleic Acids Res. 2014. PMID: 24848018 Free PMC article. Review. - The autosomal dominant spinocerebellar ataxias: emerging mechanistic themes suggest pervasive Purkinje cell vulnerability.
Hekman KE, Gomez CM. Hekman KE, et al. J Neurol Neurosurg Psychiatry. 2015 May;86(5):554-61. doi: 10.1136/jnnp-2014-308421. Epub 2014 Aug 18. J Neurol Neurosurg Psychiatry. 2015. PMID: 25136055 Free PMC article. Review. - RNA-binding proteins implicated in neurodegenerative diseases.
Cookson MR. Cookson MR. Wiley Interdiscip Rev RNA. 2017 Jan;8(1):10.1002/wrna.1397. doi: 10.1002/wrna.1397. Epub 2016 Sep 23. Wiley Interdiscip Rev RNA. 2017. PMID: 27659605 Free PMC article. Review.
References
- Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi:10.1146/annurev.neuro.29.051605.113042. - DOI - PubMed
- Batra R., Charizanis K., Swanson M.S. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–R82. doi:10.1093/hmg/ddq132. - DOI - PMC - PubMed
- Nelson D.L., Orr H.T., Warren S.T. The unstable repeats—three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi:10.1016/j.neuron.2013.02.022. - DOI - PMC - PubMed
- Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi:10.1146/annurev.neuro.29.051605.113014. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 NS058901/NS/NINDS NIH HHS/United States
- R01 NS040389/NS/NINDS NIH HHS/United States
- P01NS058901/NS/NINDS NIH HHS/United States
- R01NS040389/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous