Active DNA demethylation in post-mitotic neurons: a reason for optimism - PubMed (original) (raw)
Review
Active DNA demethylation in post-mitotic neurons: a reason for optimism
David P Gavin et al. Neuropharmacology. 2013 Dec.
Abstract
Over the last several years proteins involved in base excision repair (BER) have been implicated in active DNA demethylation. We review the literature supporting BER as a means of active DNA demethylation, and explain how the various components function and cooperate to remove the potentially most enduring means of epigenetic gene regulation. Recent evidence indicates that the same pathways implicated during periods of widespread DNA demethylation, such as the erasure of methyl marks in the paternal pronucleus soon after fertilization, are operational in post-mitotic neurons. Neuronal functional identities, defined here as the result of a combination of neuronal subtype, location, and synaptic connections are largely maintained through DNA methylation. Chronic mental illnesses, such as schizophrenia, may be the result of both altered neurotransmitter levels and neurons that have assumed dysfunctional neuronal identities. A limitation of most current psychopharmacological agents is their focus on the former, while not addressing the more profound latter pathophysiological process. Previously, it was believed that active DNA demethylation in post-mitotic neurons was rare if not impossible. If this were the case, then reversing the factors that maintain neuronal identity, would be highly unlikely. The emergence of an active DNA demethylation pathway in the brain is a reason for great optimism in psychiatry as it provides a means by which previously pathological neurons may be reprogrammed to serve a more favorable role. Agents targeting epigenetic processes have shown much promise in this regard, and may lead to substantial gains over traditional pharmacological approaches.
Keywords: 5-Hydroxymethylcytosine; 5-Methylcytosine; 5-carboxylcytosine; 5-formylcytosine; 5-hydroxymethylcytosine; 5-hydroxymethyluracil; 5-methylcytosine; 5CaC; 5FC; 5HMC; 5HMU; 5MC; AICDA; APE1; APOBEC; BDNF; BER; COMT; CpG; CpG methylation; DNA methyltransferase; DNMT; ECS; FGF; G; GADD45; H3K27; H3K9me2; HDAC; KCl; LSD1; MBD; MeCP2; N-methyl d-aspartate receptor subtype 2B; N-methyl-d-aspartic acid; NER; NMDA; NR2B; PARP1; PFC; PGC; PP1; SMUG1; T; TDG; TET; X-ray repair cross-complementing protein-1; XRCC1; activation-induced cytidine deaminase; apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; apurinic/apyrimidinic (AP) endonuclease; base excision repair; brain-derived neurotrophic factor; catechol-O-methyltransferase; cytosine–phosphate–guanine; dimethylated lysine 9 of histone 3; electroconvulsive seizure; fibroblast growth factor; growth arrest and DNA damage; guanine; histone deacetylase; lysine 27 of histone 3; lysine-specific demethylase-1; methyl CpG binding protein 2; methyl-CpG binding domain; nucleotide excision repair; poly ADP ribose polymerase-1; potassium chloride; prefrontal cortex; primordial germ cells; protein phosphatase 1; single-strand selective monofunctional uracil-DNA glycosylase 1; ten-eleven translocation; thymidine; thymine-DNA glycosylase.
Published by Elsevier Ltd.
Figures
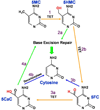
Fig 1. Molecular Changes Leading to DNA Demethylation
1) In an oxidation reaction catalyzed by ten-eleven translocation (TET) enzymes 5-methylcytosine (5MC) is converted into 5-hydroxymethylcytosine (5HMC). 2a) Through a process involving deamination followed by base excision repair 5HMC is removed and replaced by an unmethylated cytosine. 2b) Alternatively, 5HMC may be converted into 5-formylcytosine (5FC) in another oxidation reaction catalyzed by TET. 3a) 5FC can be converted into 5-carboxylcytosine (5CaC) in another TET-catalyzed oxidation reaction. 3b) 5FC hypothetically could be converted directly into unmethylated cytosine involving the expulsion of formic acid. 4a) 5CaC can be removed via base excision repair. 4b) 5CaC hypothetically could be converted directly into unmethylated cytosine in a reaction catalyzed by an as yet unidentified decarboxylase.
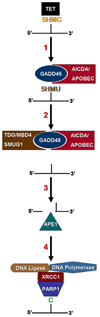
Fig 2. Base Excision Repair DNA Demethylation Pathway
1. Following the TET catalyzed hydroxylation of 5-methylcytosine to 5-hydroxymethylcytosine (5HMC), 5HMC is deaminated by a Growth Arrest and DNA Damage (GADD45) protein recruited cytidine deaminase, such as AICDA or an APOBEC forming 5-hydroxymethyluracil (5HMU). 2. GADD45 proteins also recruit a thymidine or uracil glycosylase, such as TDG, MBD4, or SMUG1, which then removes the 5HMU base. 3. APE1 recognizes the now apyrimidinic site and creates a strand break 5’ to the missing base. 4. PARP1 is strongly induced by the resultant strand break. PARP1 dimerizes at the break and also adds Poly(ADP-Ribose) (PAR) groups to proteins such as scaffold protein XRCC1. The addition of PAR groups facilitates protein-protein interactions between XRCC1 and DNA ligase and polymerase, which repair the break and replace the missing base.
Fig 3. Increased expression of demethylating genes in schizophrenia vs. nonpsychiatric controls in two cohorts
We re-analyzed microarray data from the National Brain Databank established at Harvard Brain Tissue Resource Center (Harvard) obtained from the website (
http://national\_databank.mclean.harvard.edu/brainbank/Main
) and Scripps Research Institute (Scripps) for genes involved in DNA demethylation. The Harvard study utilized the Affymetrix HG-U133A gene chip. Experimental result files had been generated using Affymetrix's MAS software. These include prefrontal cortical (PFC) samples from 25 normal control subjects and 19 with schizophrenia. Demographic details have been previously published (Sharma et al., 2008a). The result files were then merged using Affymetrix Expression Console Software with the annotation file and the annotated log2 results exported as a text file for third-party downstream analysis. The Scripps study utilized the Human Genome U133 Plus 2.0 array. We reanalyzed this post mortem expression data obtained from the Gene expression Omnibus (GEO accession GSE21138) based on diagnosis. The database includes PFC samples from 59 total subjects, 29 normal controls and 30 patients with schizophrenia. Demographic details have been previously published (Narayan et al., 2008). Expression was converted from log2 expression to linear expression as fold average of normal controls. Independent t-tests were used for group comparisons. All p-values are two-tailed. A. AICDA, APOBEC3A, and APOBEC3F are significantly increased in the Harvard study (*P < 0.05). B. APOBEC3B and APOBEC3G are increased in the Scripps dataset (*P < 0.05; **P < 0.01). C & D. GADD45B and GADD45G are increased in both datasets. GADD45B has previously been shown to be elevated in the Stanley Medical research cohort as well (
https://www.stanleygenomics.org/
;(Gavin et al., 2012), while GADD45G has also been previously reported to be elevated in schizophrenia (Arion et al., 2007) (*P < 0.05). E. TET3 is increased in the Harvard dataset (*P < 0.05). F. MBD4 is significantly increased in the Scripps cohort. An increase in MBD4 in schizophrenia has previously been reported by Benes et al. (2006). Despite the consistent increase in DNA demethylating factors we do find a decrease in SMUG1 (*P < 0.05; ***P < 0.001). Mean+SEM.
Fig 4. Euchromatization
1. The first step in the progression from restrictive heterochromatin to transcriptionally permissive euchromatin begins with the removal of histone marks such as, dimethylated lysine 9 of histone 3 (H3K9me2), and H3K9 acetylation. Restrictive histone modifications bar the DNA demethylation complex from accessing the underlying cytosine. 2. H3K9 demethylation and acetylation allow for transcription factor binding as well as the binding of the mediators of DNA demethylation. Histone acetylation and the DNA demethylation process promote the addition of Poly(ADP-Ribose) polymers (PAR) to histone lysine residues such as H3K27. 3. Once DNA is demethylated a H3K4 methyltransferase can bind. Methylated H3K4 prevents DNMT binding thereby maintaining the unmethylated DNA state.
Similar articles
- Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase.
van der Wijst MG, Venkiteswaran M, Chen H, Xu GL, Plösch T, Rots MG. van der Wijst MG, et al. Epigenetics. 2015;10(8):671-6. doi: 10.1080/15592294.2015.1062204. Epigenetics. 2015. PMID: 26098813 Free PMC article. Review. - PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells.
Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, Hashimoto S, Nakamura T, Sugasawa K, Kojima N, Takada T, Okano M, Seki Y. Okashita N, et al. Development. 2014 Jan;141(2):269-80. doi: 10.1242/dev.099622. Epub 2013 Dec 11. Development. 2014. PMID: 24335252 - Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Hashimoto H, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10203-14. doi: 10.1093/nar/gks845. Epub 2012 Sep 8. Nucleic Acids Res. 2012. PMID: 22962365 Free PMC article. - Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites.
Maiti A, Drohat AC. Maiti A, et al. J Biol Chem. 2011 Oct 14;286(41):35334-35338. doi: 10.1074/jbc.C111.284620. Epub 2011 Aug 23. J Biol Chem. 2011. PMID: 21862836 Free PMC article. - DNA demethylation pathways: Additional players and regulators.
Bochtler M, Kolano A, Xu GL. Bochtler M, et al. Bioessays. 2017 Jan;39(1):1-13. doi: 10.1002/bies.201600178. Epub 2016 Nov 16. Bioessays. 2017. PMID: 27859411 Review.
Cited by
- Molecular mechanisms of synaptic remodeling in alcoholism.
Kyzar EJ, Pandey SC. Kyzar EJ, et al. Neurosci Lett. 2015 Aug 5;601:11-9. doi: 10.1016/j.neulet.2015.01.051. Epub 2015 Jan 23. Neurosci Lett. 2015. PMID: 25623036 Free PMC article. Review. - Epigenetics of Subcellular Structure Functioning in the Origin of Risk or Resilience to Comorbidity of Neuropsychiatric and Cardiometabolic Disorders.
Zapata-Martín Del Campo CM, Martínez-Rosas M, Guarner-Lans V. Zapata-Martín Del Campo CM, et al. Int J Mol Sci. 2018 May 14;19(5):1456. doi: 10.3390/ijms19051456. Int J Mol Sci. 2018. PMID: 29757967 Free PMC article. Review. - DNMT1 modulates interneuron morphology by regulating Pak6 expression through crosstalk with histone modifications.
Symmank J, Bayer C, Schmidt C, Hahn A, Pensold D, Zimmer-Bensch G. Symmank J, et al. Epigenetics. 2018;13(5):536-556. doi: 10.1080/15592294.2018.1475980. Epub 2018 Aug 7. Epigenetics. 2018. PMID: 29912614 Free PMC article. - Estradiol differentially induces progesterone receptor isoforms expression through alternative promoter regulation in a mouse embryonic hypothalamic cell line.
Vázquez-Martínez ER, Camacho-Arroyo I, Zarain-Herzberg A, Rodríguez MC, Mendoza-Garcés L, Ostrosky-Wegman P, Cerbón M. Vázquez-Martínez ER, et al. Endocrine. 2016 Jun;52(3):618-31. doi: 10.1007/s12020-015-0825-1. Epub 2015 Dec 16. Endocrine. 2016. PMID: 26676302 - Chromatin Switches during Neural Cell Differentiation and Their Dysregulation by Prenatal Alcohol Exposure.
Gavin DP, Grayson DR, Varghese SP, Guizzetti M. Gavin DP, et al. Genes (Basel). 2017 May 11;8(5):137. doi: 10.3390/genes8050137. Genes (Basel). 2017. PMID: 28492482 Free PMC article. Review.
References
- Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: A preliminary report. American Journal of Medical Genetics.Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of Psychiatric Genetics. 2005;134B:60–66. - PubMed
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Research Reviews. 2006;52:293–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous