Bacterial argonaute samples the transcriptome to identify foreign DNA - PubMed (original) (raw)
Bacterial argonaute samples the transcriptome to identify foreign DNA
Ivan Olovnikov et al. Mol Cell. 2013.
Abstract
Eukaryotic Argonautes bind small RNAs and use them as guides to find complementary RNA targets and induce gene silencing. Though homologs of eukaryotic Argonautes are present in many bacteria and archaea, their small RNA partners and functions are unknown. We found that the Argonaute of Rhodobacter sphaeroides (RsAgo) associates with 15-19 nt RNAs that correspond to the majority of transcripts. RsAgo also binds single-stranded 22-24 nt DNA molecules that are complementary to the small RNAs and enriched in sequences derived from exogenous plasmids as well as genome-encoded foreign nucleic acids such as transposons and phage genes. Expression of RsAgo in the heterologous E. coli system leads to formation of plasmid-derived small RNA and DNA and plasmid degradation. In a R. sphaeroides mutant lacking RsAgo, expression of plasmid-encoded genes is elevated. Our results indicate that RNAi-related processes found in eukaryotes are also conserved in bacteria and target foreign nucleic acids.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
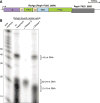
Figure 1. R. sphaeroides Argonaute protein associates with small RNA and small DNA molecules
(A) R. sphaeroides Argonaute protein and its operon structure. Gene Rsph17025_3695 encodes a predicted nuclease and overlaps with the last 4 nt of the RsAgo coding sequence. PAZ, MID and PIWI are conserved domains shared by all Argonautes, while N (N-terminal), L1 (linker 1) and L2 (linker 2) are not conserved. (B) Purified RsAgo is associated with small RNA (15-19 nt) and small DNA (22-24 nt) species. RsAgo-associated nucleic acids were 5′-radioactively labeled, treated with RNase A or DNase I and resolved on a 15% urea PAGE. The ∼45 nt band is composed primarily of tRNAmet 3′ fragments. Densitometric measurement of three independent RsAgo samples indicates that the ratio of small RNA to small DNA in RsAgo complexes is 2.12±0.66.
Figure 2. Analysis of RsAgo-associated small RNA
(A) Read length distribution of sequenced small RNA and small DNA associated with RsAgo. Only sequences that could be mapped to the genome or the expression plasmid were considered. (B) Nucleotide bias in small RNA extracted from purified RsAgo. 16-19 nt reads were trimmed from the 3′end to 16 nt and analyzed with WebLogo. (C) Length profile of total small RNAs extracted from two strains of R. sphaeroides - one encoding Ago (17025) and one not (17029) - carrying either an empty plasmid or a plasmid expressing RsAgo. (D) Bias for the uridine residue in position 1 in libraries shown in panel C. 61% of reads in total small RNA from wild type strain 17025 lacking expression plasmid had uridine in position 1.
Figure 3. RsAgo-associated small RNAs represent a broad sample of the transcriptome
(A) Distribution of unique small RNA reads from purified RsAgo over the six chromosomes of R. sphaeroides strain 25 plotted as a fraction of raw read numbers and read numbers normalized to chromosome length. (B) Annotation of RsAgo-associated small RNAs. Genes on the expression plasmid were included in the analysis. (C) Density of cloned RsAgo-associated small RNAs along chromosome 2 of R. sphaeroides strain 25. The graph includes reads that can be mapped without mismatch to multiple genomic positions. Note that the large number of reads mapping to the RsAgo gene is likely originating from the expression plasmid. In the top graph each genomic position corresponding to a cloned small RNA is given equal weight; on the bottom graph each read is normalized for the number of times it maps to the genome, resulting in proportionally less signal for reads that map many times such small RNA matching IS4 transposon. (D) The correlation between long and small RNA abundance for R. sphaeroides genes. rRNA-depleted long RNA library was prepared from strain 25 containing the pSRKKm-RsAgo plasmid. (E)R. sphaeroides genes are sorted by their enrichment for small RNAs relative to long transcripts as measured by RNA-Seq. Shown are fold enrichment (positive values) or depletion (negative values) of small to long RNA normalized to the mean small RNA/long RNA ratio for all genes. The 100 genes most strongly depleted in small RNA (right side of the distribution) are enriched in non-coding RNA relative to the rest (12% compared to 1.2%).
Figure 4. Analysis of RsAgo-associated small DNA
(A) Distribution of unique small DNA reads from purified RsAgo over the six chromosomes of R. sphaeroides strain 25 plotted as a fraction of raw read numbers and read numbers normalized to chromosome length. (B) Annotation of RsAgo-associated small DNA sequences. (C) The profile of RsAgo-associated small RNA and DNA over the expression plasmid pSRKKm-RsAgo. Reads that mapped to the plus or minus strand are shown above and below the axis, respectively. (D) Strand bias of RsAgo small RNAs and DNAs mapped to genes encoded on the expression plasmid pSRKKm-RsAgo. (E) Relative distance between ends of small RNAs and DNAs mapped to opposite genomic strands of plasmid pSRKKm-RsAgo. Graphs indicate that RNA and DNA have a high tendency to map as pairs with the DNA molecule protruding 3 nt on each side. Note that this distance is more rigid on the left side (5′ end of RNA and 3′ end of DNA) compared to the right side (3′ end of RNA and 5′ end of DNA) (F) Nucleotide bias in small DNA aligned by the 3′ end and analyzed with WebLogo. (G) Model of small diRNA-riDNA pairs.
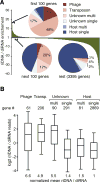
Figure 5. Distribution of diRNA and riDNA over R. sphaeroides genes
(A) Fold enrichment (positive values) or depletion (negative values) of riDNA/diRNA ratio normalized to the mean riDNA/diRNA ratio for all genes. Genes are sorted by their enrichment for riDNA relative to diRNA: riDNA-rich genes are on the left, depleted on the right side of the graph. The frequencies of six different gene classes of the R. sphaeroides strain 25 genome (single and multi-copy host genes, single- and multi-copy genes of unknown origin and phage- and transposon-related genes) were analyzed for the 100 most DNA-rich genes, the next 100 genes in the distribution and the rest of the distribution (3395 genes). For a similar analysis of small DNA reads normalized to the genomic copy number see Fig. S5C. (B) Box plots of riDNA to diRNA ratios for the same gene classes as in panel A. Number of genes in each class is shown above the plot. The box represents the 25th, 50th (the inner line) and the 75th percentiles of the distribution; whiskers are at the 5th and 95th percentile. The mean of the ratio of riDNA to diRNA was calculated for each gene class and normalized to that of host single-copy genes. The same plot with DNA read numbers divided by the number of genome mappings (copy number) in shown on Fig. S5D.
Figure 6. The effects of RsAgo expression in_E. coli_
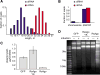
(A) Read length distribution of sequenced small RNA and small DNA associated with RsAgo expressed in E. coli strain BL21(DE3). Only sequences that could be mapped to the genome and the expression plasmid were considered. (B) Fractions of small RNA and small DNA reads from RsAgo purified from E. coli BL21(DE3) that map to the host chromosome and expression plasmid (C) Plasmid yields from E. coli BL21(DE3) cells after induction of expression of GFP, wild-type RsAgo and RsAgo mutant impaired in small RNA binding (RsAgo-YK). Shown is the mean plasmid yield (+/- standard deviation) per cell density unit (OD600). (D) RsAgo expression in E. coli causes degradation of plasmid DNA. Equal amounts of plasmid DNA from the experiment shown on panel C were resolved on agarose gel and stained with SYBR Gold. The expression of proteins was induced with IPTG and shown on Fig. S6.
Figure 7. The effects of RsAgo mutation in_R. sphaeroides_
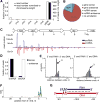
(A) Read length distribution of sequenced total small RNA (13 – 30 nt) from wild-type and RsAgo mutant R. sphaeroides strain 25 cells. Only sequences that could be mapped to the genome were considered. (B) Comparison of expression of plasmid-encoded genes in wild-type and RsAgo mutant R. sphaeroides strain 25. The luciferase (Fluc) and lacI genes are encoded on the plasmid pSRKTc-Fluc. Firefly luciferase activity was quantified by measuring luminescence. Shown is normalized mean ratio of luminescence in induced to non-induced cells +/- SD (n=3). Fluc and lacI mRNA was quantified by RT-qPCR and normalized to the average mRNA level of three host genes (normalized mean +/- SD, n=3). pSRKTc-Fluc plasmid copy number was determined by qPCR. (C) The model of RsAgo function and biogenesis of diRNA and riDNA. diRNAs are generated from mRNAs (a-c) or products of their degradation, while structural non-coding RNA (d) is not efficiently processed into small RNA. RsAgo loaded with cognate diRNA (c) recognizes plasmid DNA followed by excision of 22-24 nt single-stranded target DNA fragment leading to damage of DNA template. Binding of plasmid DNA by RsAgo/diRNA complex might also interfere with transcription by RNA polymerase prior to DNA excision. Excised riDNA forms a duplex with guide diRNA in the RsAgo complex and does not play further role in repression. Alternatively, riDNA is loaded into a new RsAgo complex and directs it to the plasmid transcript to induce post-transcriptional repression.
Comment in
- Bacterial physiology: bacterial argonaute sets sail.
Molloy S. Molloy S. Nat Rev Microbiol. 2013 Nov;11(11):743. doi: 10.1038/nrmicro3150. Nat Rev Microbiol. 2013. PMID: 24129508 No abstract available. - Biochemistry. A bacterial seek-and-destroy system for foreign DNA.
Vogel J. Vogel J. Science. 2014 May 30;344(6187):972-3. doi: 10.1126/science.1252962. Science. 2014. PMID: 24876480 No abstract available.
Similar articles
- Recognition of double-stranded DNA by the Rhodobacter sphaeroides Argonaute protein.
Lisitskaya L, Petushkov I, Esyunina D, Aravin A, Kulbachinskiy A. Lisitskaya L, et al. Biochem Biophys Res Commun. 2020 Dec 17;533(4):1484-1489. doi: 10.1016/j.bbrc.2020.10.051. Epub 2020 Oct 24. Biochem Biophys Res Commun. 2020. PMID: 33333714 - Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.
Miyoshi T, Ito K, Murakami R, Uchiumi T. Miyoshi T, et al. Nat Commun. 2016 Jun 21;7:11846. doi: 10.1038/ncomms11846. Nat Commun. 2016. PMID: 27325485 Free PMC article. - Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Liu Y, Esyunina D, Olovnikov I, Teplova M, Kulbachinskiy A, Aravin AA, Patel DJ. Liu Y, et al. Cell Rep. 2018 Jul 10;24(2):453-462. doi: 10.1016/j.celrep.2018.06.021. Cell Rep. 2018. PMID: 29996105 Free PMC article. - Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes.
Olina AV, Kulbachinskiy AV, Aravin AA, Esyunina DM. Olina AV, et al. Biochemistry (Mosc). 2018 May;83(5):483-497. doi: 10.1134/S0006297918050024. Biochemistry (Mosc). 2018. PMID: 29738683 Review. - Argonaute proteins confer immunity in all domains of life.
Bobadilla Ugarte P, Barendse P, Swarts DC. Bobadilla Ugarte P, et al. Curr Opin Microbiol. 2023 Aug;74:102313. doi: 10.1016/j.mib.2023.102313. Epub 2023 Apr 4. Curr Opin Microbiol. 2023. PMID: 37023508 Review.
Cited by
- Small-RNA loading licenses Argonaute for assembly into a transcriptional silencing complex.
Holoch D, Moazed D. Holoch D, et al. Nat Struct Mol Biol. 2015 Apr;22(4):328-35. doi: 10.1038/nsmb.2979. Epub 2015 Mar 2. Nat Struct Mol Biol. 2015. PMID: 25730778 Free PMC article. - Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion.
Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V. Zaremba M, et al. Nat Microbiol. 2022 Nov;7(11):1857-1869. doi: 10.1038/s41564-022-01239-0. Epub 2022 Oct 3. Nat Microbiol. 2022. PMID: 36192537 - Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Gao X, Shang K, Zhu K, Wang L, Mu Z, Fu X, Yu X, Qin B, Zhu H, Ding W, Cui S. Gao X, et al. Nature. 2024 Jan;625(7996):822-831. doi: 10.1038/s41586-023-06665-6. Epub 2023 Oct 2. Nature. 2024. PMID: 37783228 - Catalytically inactive long prokaryotic Argonaute systems employ distinct effectors to confer immunity via abortive infection.
Song X, Lei S, Liu S, Liu Y, Fu P, Zeng Z, Yang K, Chen Y, Li M, She Q, Han W. Song X, et al. Nat Commun. 2023 Nov 1;14(1):6970. doi: 10.1038/s41467-023-42793-3. Nat Commun. 2023. PMID: 37914725 Free PMC article. - RNA damage in biological conflicts and the diversity of responding RNA repair systems.
Burroughs AM, Aravind L. Burroughs AM, et al. Nucleic Acids Res. 2016 Oct 14;44(18):8525-8555. doi: 10.1093/nar/gkw722. Epub 2016 Aug 17. Nucleic Acids Res. 2016. PMID: 27536007 Free PMC article. Review.
References
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials