New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion - PubMed (original) (raw)
New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion
Christiane Margue et al. PLoS One. 2013.
Abstract
The non-coding microRNAs (miRNA) have tissue- and disease-specific expression patterns. They down-regulate target mRNAs, which likely impacts on most fundamental cellular processes. Differential expression patterns of miRNAs are currently being exploited for identification of biomarkers for early disease diagnosis, prediction of progression for melanoma and other cancers and as promising drug targets, since they can easily be inhibited or replaced in a given cellular context. Before successfully manipulating miRNAs in clinical settings, their precise expression levels, endogenous functions and thus their target genes have to be determined. MiR-211, a melanocyte lineage-specific small non-coding miRNA, is located in an intron of TRPM1, a target gene of the microphtalmia-associated transcription factor (MITF). By transcriptionally up-regulating TRPM1, MITF, which is critical for both melanocyte differentiation and survival and for melanoma progression, indirectly drives the expression of miR-211. Expression of this miRNA is often reduced in melanoma samples. Here, we investigated functional roles of miR-211 by identifying and studying new target genes. We show that MITF-correlated miR-211 expression levels are mostly but not always reduced in a panel of 11 melanoma cell lines and in primary and metastatic melanoma compared to normal melanocytes and nevi, respectively. MiR-211 itself only marginally impacted on cell invasion and migration, while perturbation of some new miR-211 target genes, such as AP1S2, SOX11, IGFBP5, and SERINC3 significantly increased invasion. These results and the variable expression levels of miR-211 raise serious doubts on the value of miR-211 as a melanoma tumor-suppressing miRNA and/or as a biomarker for melanoma.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
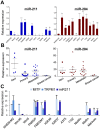
Figure 1. Expression profiling of miR-211 and co-expressed proteins.
(A) RNA of primary melanocytes (NHEM-M2) and 11 different melanoma cell lines was analyzed for relative miR-211 (blue) and miR-204 (red) expression levels by qPCR. Statistical significance was assessed with ANOVA (repeated measures) followed by a Dunnett Post-Hoc multiple comparison test. P values of <0.05 (*), <0.01 (**) and <0.001 (***) were considered significant. (B) RNA of FFPE patient samples from 4 nevi, 9 primary and 12 metastatic melanoma samples and 2 breast cancer samples were analyzed as above. (C) Co-expression of MITF, TRPM1 and the intronic miR-211 was confirmed in the same cell lines and statistical significance was tested as in (A). Except for FFPE patient samples, all experiments were performed at least in biological triplicates.
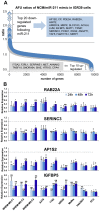
Figure 2. Identification of miR-211 target genes.
(A) To identify new miR-211 target genes, miR-211-negative IGR39 melanoma cells were transfected with 5 nM miR-211 mimic or negative control (NCM) for 48 h. RNAs from two independent samples were extracted and analyzed separately by Affymetrix mRNA arrays. The scatter plots show ratios of normalized and averaged arbitrary fluorescence units (AFU) from two independent arrays between NCM- and mimic-treated cells for all mRNAs. Top-regulated genes are listed in grey boxes. (B) Expression levels (qPCR) of 4 selected and putative miR-211 targets RAB22A, SERINC3, AP1S2 and IGFBP5 after 5 nM miR-211 mimic or NCM (or 10 nM for primary melanocytes NHEM) treatment for 24, 48 and 72 h +/− SEM are shown. Levels of NCM-treated cells were set to 1. Significance was tested with a t-test and p values are as described in Figure 1.
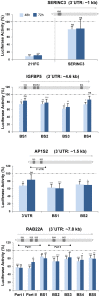
Figure 3. Luciferase reporter gene assays confirm new direct targets of miR-211 in melanoma cells.
A luciferase reporter vector containing indicated stretches of the target 3′UTR or single binding sites (BS) for miR-211 (depicted by schemes above the graphs) were transfected together with 5 nM miR-211 mimic or NCM into A375 melanoma cells. After 48 and 72 h, luciferase activity was measured. The full complementary (FC) sequence of miR-211 was cloned into the luciferase vector and served as a positive control. Shown are ratios of mimic/NCM-treated cells. Activity of NCM-treated cells was set to 1 with bars representing the average of at least 3 biological replicates per time point, +/− SEM. Significance was tested by a paired t-test with p values as described above.
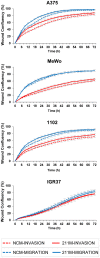
Figure 4. MiR-211 has no significant effects on melanoma cell invasion and migration.
Four melanoma cell lines were transfected with miR-211 mimic (solid lines) or NCM (dotted lines); successful mimic transfection was confirmed by qPCR in all samples (Figure S5). After 24 h, a scratch/wound assay was performed and cells were grown for up to 72 h. To study invasion (red lines), cells were covered with a collagen matrix after wound scratching; to study migration (blue lines), cells were grown in wells without a collagen matrix containing only normal growth medium. Invasion and migration were monitored by measuring wound closure every 3 h for a total of 72 h with the Incucyte LiveCell Imaging System (Essen Bioscience). Basal levels of miR-211 in these cell lines are shown in Figure 1. The graphs depict averages from two biological replicates each including at least quadruplicate samples +/−SEM. In cells without endogenous miR-211 (MeWo, 1102, and A375), the mimic treatment caused a very small but reproducible increase in invasion; in IGR37 (high endogenous miR-211), a further increase of miR-211 by transfection did not affect invasion/migration at all.
Figure 5. siRNA-mediated down-regulation of new miR-211 targets has an impact on melanoma cell invasion and migration.
A375 cells were transfected with siRNAs directed against selected miR-211 targets. After 24 h, a scratch/wound assay was performed as described in Figure 4. To ensure efficient siRNA-mediated down-regulation of target mRNAs, qPCR was performed on total RNA (extracted from 4 pooled wells for each treatment) at 24, 48 and 72 h after transfection in the invasion and migration wells (small inlets, upper left corners). Representative graphs of four biological replicate experiments are shown. Error bars show STD from at least 4 technical replicates for each measurement.
Similar articles
- miR-211 and MITF modulation by Bcl-2 protein in melanoma cells.
De Luca T, Pelosi A, Trisciuoglio D, D'Aguanno S, Desideri M, Farini V, Di Martile M, Bellei B, Tupone MG, Candiloro A, Regazzo G, Rizzo MG, Del Bufalo D. De Luca T, et al. Mol Carcinog. 2016 Dec;55(12):2304-2312. doi: 10.1002/mc.22437. Epub 2015 Nov 24. Mol Carcinog. 2016. PMID: 26599548 - Signatures of microRNAs and selected microRNA target genes in human melanoma.
Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Philippidou D, et al. Cancer Res. 2010 May 15;70(10):4163-73. doi: 10.1158/0008-5472.CAN-09-4512. Epub 2010 May 4. Cancer Res. 2010. PMID: 20442294 - The roles of microphthalmia-associated transcription factor and pigmentation in melanoma.
Hsiao JJ, Fisher DE. Hsiao JJ, et al. Arch Biochem Biophys. 2014 Dec 1;563:28-34. doi: 10.1016/j.abb.2014.07.019. Epub 2014 Aug 9. Arch Biochem Biophys. 2014. PMID: 25111671 Free PMC article. Review. - Microphthalmia-associated transcription factor expression levels in melanoma cells contribute to cell invasion and proliferation.
Vachtenheim J, Ondrušová L. Vachtenheim J, et al. Exp Dermatol. 2015 Jul;24(7):481-4. doi: 10.1111/exd.12724. Exp Dermatol. 2015. PMID: 25866058 Review.
Cited by
- LINC00518: a key player in tumor progression and clinical outcomes.
Yi Q, Zhu G, Zhu W, Wang J, Ouyang X, Yang K, Zhong J. Yi Q, et al. Front Immunol. 2024 Jul 23;15:1419576. doi: 10.3389/fimmu.2024.1419576. eCollection 2024. Front Immunol. 2024. PMID: 39108268 Free PMC article. Review. - Identification of Keratinocyte Differentiation-Involved Genes for Metastatic Melanoma by Gene Expression Profiles.
Li K, Guo S, Tong S, Sun Q, Jin S, Qi B, Shao Y, Xu N. Li K, et al. Comput Math Methods Med. 2021 Dec 28;2021:9652768. doi: 10.1155/2021/9652768. eCollection 2021. Comput Math Methods Med. 2021. PMID: 35003328 Free PMC article. - Transferring intercellular signals and traits between cancer cells: extracellular vesicles as "homing pigeons".
Cesi G, Walbrecq G, Margue C, Kreis S. Cesi G, et al. Cell Commun Signal. 2016 Jun 10;14(1):13. doi: 10.1186/s12964-016-0136-z. Cell Commun Signal. 2016. PMID: 27282631 Free PMC article. Review. - MicroRNA-211 Loss Promotes Metabolic Vulnerability and BRAF Inhibitor Sensitivity in Melanoma.
Sahoo A, Sahoo SK, Joshi P, Lee B, Perera RJ. Sahoo A, et al. J Invest Dermatol. 2019 Jan;139(1):167-176. doi: 10.1016/j.jid.2018.06.189. Epub 2018 Aug 1. J Invest Dermatol. 2019. PMID: 30076926 Free PMC article. - The miRNAs Role in Melanoma and in Its Resistance to Therapy.
Varrone F, Caputo E. Varrone F, et al. Int J Mol Sci. 2020 Jan 29;21(3):878. doi: 10.3390/ijms21030878. Int J Mol Sci. 2020. PMID: 32013263 Free PMC article. Review.
References
- Weiland M, Gao XH, Zhou L, Mi QS (2012) Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol 9: 850–859. - PubMed
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, et al. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was funded by a research grant from the University of Luxembourg (F1R_LSC-PUL-09MIRN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous