PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage - PubMed (original) (raw)
PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage
Stuart L Rulten et al. Nucleic Acids Res. 2014 Jan.
Abstract
Amyotrophic lateral sclerosis (ALS) is associated with progressive degeneration of motor neurons. Several of the genes associated with this disease encode proteins involved in RNA processing, including fused-in-sarcoma/translocated-in-sarcoma (FUS/TLS). FUS is a member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family of proteins that bind thousands of pre-mRNAs and can regulate their splicing. Here, we have examined the possibility that FUS is also a component of the cellular response to DNA damage. We show that both GFP-tagged and endogenous FUS re-localize to sites of oxidative DNA damage induced by UVA laser, and that FUS recruitment is greatly reduced or ablated by an inhibitor of poly (ADP-ribose) polymerase activity. Consistent with this, we show that recombinant FUS binds directly to poly (ADP-ribose) in vitro, and that both GFP-tagged and endogenous FUS fail to accumulate at sites of UVA laser induced damage in cells lacking poly (ADP-ribose) polymerase-1. Finally, we show that GFP-FUS(R521G), harbouring a mutation that is associated with ALS, exhibits reduced ability to accumulate at sites of UVA laser-induced DNA damage. Together, these data suggest that FUS is a component of the cellular response to DNA damage, and that defects in this response may contribute to ALS.
Figures
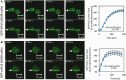
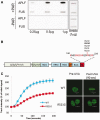
Figure 1.
GFP-tagged FUS/TLS accumulates at sites of UVA laser-induced oxidative chromosomal damage. (A) Recruitment of eGFP-hFUS to sites of laser damage. Transiently transfected A549 cells were subjected to UVA laser microirradiation along the line indicated. Images were taken at the times (seconds) shown after microirradiation. The graph shows the average GFP fluorescence across six individual experiments and over 100 cells ± SEM. (B) Recruitment of eGFP-mFUS to sites of laser damage. Experiments were carried as described in (A). Graph shows the mean of three independent experiments.
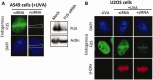
Figure 2.
FUS/TLS accumulates at sites of UVA laser-induced oxidative chromosomal damage. (A) A549 cells mock-treated (‘−siRNA’) or pretreated with FUS siRNA (‘+siRNA’) were microirradiated, fixed 2 min later and immunostained for endogenous FUS with anti-FUS antibody (top two panels). A western blot confirming siRNA-mediated knockdown is shown (right). The dotted box in the ‘+siRNA’ samples denotes the position of the UVA laser track. (B) U2OS cells were treated as described above and immunostained with anti-FUS (middle row), or anti-γH2Ax antibody (bottom row) as a marker of DNA breaks.
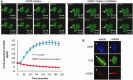
Figure 3.
FUS/TLS accumulation at sites of UVA laser-induced damage is dependent on PAR synthesis. (A) Human A549 cells were transfected with GFP-hFUS and microirradiated with UVA. Cells were pretreated with vehicle (DMSO) or 500 nM KU58948 1 h before microirradiation. A representative experiment is shown with quantification of GFP-FUS recruitment (mean ± SEM > 30 cells) plotted graphically (bottom left). (B) U2OS cells mock-treated or pretreated with 1 µM KU58948 (PARPi) were microirradiated as described above, fixed and immunolabelled for endogenous FUS (middle row) and γH2Ax (bottom).
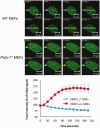
Figure 4.
FUS/TLS accumulation at sites of UVA laser-induced damage is dependent on PARP-1. Parp-1+/+ (WT) and Parp-1−/−MEFs were transiently transfected with GFP-hFUS and microirradiated with a UVA laser. Representative images are presented, with quantification of GFP-FUS recruitment (mean ± SEM > 20 cells) plotted graphically (bottom).
Figure 5.
FUS/TLS binds directly to PAR, and a mutation associated with ALS disrupts FUS/TLS recruitment at sites of DNA damage. (A) FUS interacts directly with PAR. The indicated amounts of recombinant hFUS or hAPLF were slot blotted onto nitrocellulose membrane and then incubated with (‘+PAR’) or without (−PAR) poly (ADP-ribose). Bound PAR was detected by western blotting. (B) Domain structure of FUS/TLS, showing the glutamine/glycine/serine/tyrosine-rich (Q/G/S/Y-rich), glycine-rich (Gly-rich), arginine/glycine-rich (RGG), RNA-binding RRM (RRM), zinc finger (ZF) and nuclear localization (NL) domains. ALS mutations associated with R521 in the NL domain are shown. (C) A549 cells transiently transfected with GFP expression construct encoding either wild type (WT) GFP-hFUS or GFP-hFUSR521G were microirradiated with UVA laser and images collected at the indicated times following irradiation. A representative image taken at 90 s following irradiation is shown (right).
Similar articles
- FUS RRM regulates poly(ADP-ribose) levels after transcriptional arrest and PARP-1 activation on DNA damage.
Mamontova EM, Clément MJ, Sukhanova MV, Joshi V, Bouhss A, Rengifo-Gonzalez JC, Desforges B, Hamon L, Lavrik OI, Pastré D. Mamontova EM, et al. Cell Rep. 2023 Oct 31;42(10):113199. doi: 10.1016/j.celrep.2023.113199. Epub 2023 Oct 5. Cell Rep. 2023. PMID: 37804508 - The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage.
Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. Mastrocola AS, et al. J Biol Chem. 2013 Aug 23;288(34):24731-41. doi: 10.1074/jbc.M113.497974. Epub 2013 Jul 5. J Biol Chem. 2013. PMID: 23833192 Free PMC article. - Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in Amyotrophic Lateral Sclerosis.
Wang H, Guo W, Mitra J, Hegde PM, Vandoorne T, Eckelmann BJ, Mitra S, Tomkinson AE, Van Den Bosch L, Hegde ML. Wang H, et al. Nat Commun. 2018 Sep 11;9(1):3683. doi: 10.1038/s41467-018-06111-6. Nat Commun. 2018. PMID: 30206235 Free PMC article. - Fused in Sarcoma (FUS) in DNA Repair: Tango with Poly(ADP-ribose) Polymerase 1 and Compartmentalisation of Damaged DNA.
Sukhanova MV, Singatulina AS, Pastré D, Lavrik OI. Sukhanova MV, et al. Int J Mol Sci. 2020 Sep 24;21(19):7020. doi: 10.3390/ijms21197020. Int J Mol Sci. 2020. PMID: 32987654 Free PMC article. Review. - TDP-43 and FUS/TLS: sending a complex message about messenger RNA in amyotrophic lateral sclerosis?
Strong MJ, Volkening K. Strong MJ, et al. FEBS J. 2011 Oct;278(19):3569-77. doi: 10.1111/j.1742-4658.2011.08277.x. Epub 2011 Sep 6. FEBS J. 2011. PMID: 21810174 Review.
Cited by
- Nucleo-cytoplasmic transport defects and protein aggregates in neurodegeneration.
Bitetto G, Di Fonzo A. Bitetto G, et al. Transl Neurodegener. 2020 Jul 3;9(1):25. doi: 10.1186/s40035-020-00205-2. Transl Neurodegener. 2020. PMID: 32616075 Free PMC article. Review. - DNA damage and regulation of protein homeostasis.
Paull TT. Paull TT. DNA Repair (Amst). 2021 Sep;105:103155. doi: 10.1016/j.dnarep.2021.103155. Epub 2021 Jun 8. DNA Repair (Amst). 2021. PMID: 34116476 Free PMC article. Review. - Pathological Involvement of Protein Phase Separation and Aggregation in Neurodegenerative Diseases.
Wu Y, Ma B, Liu C, Li D, Sui G. Wu Y, et al. Int J Mol Sci. 2024 Sep 23;25(18):10187. doi: 10.3390/ijms251810187. Int J Mol Sci. 2024. PMID: 39337671 Free PMC article. Review. - Poly-ADP ribosylation in DNA damage response and cancer therapy.
Hou WH, Chen SH, Yu X. Hou WH, et al. Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:82-91. doi: 10.1016/j.mrrev.2017.09.004. Epub 2017 Sep 20. Mutat Res Rev Mutat Res. 2019. PMID: 31395352 Free PMC article. Review. - Cell-Type-Dependent Recruitment Dynamics of FUS Protein at Laser-Induced DNA Damage Sites.
Niu Y, Pal A, Szewczyk B, Japtok J, Naumann M, Glaß H, Hermann A. Niu Y, et al. Int J Mol Sci. 2024 Mar 20;25(6):3526. doi: 10.3390/ijms25063526. Int J Mol Sci. 2024. PMID: 38542501 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous