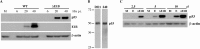Extensive post-translational modification of active and inactivated forms of endogenous p53 - PubMed (original) (raw)
Extensive post-translational modification of active and inactivated forms of endogenous p53
Caroline J DeHart et al. Mol Cell Proteomics. 2014 Jan.
Abstract
The p53 tumor suppressor protein accumulates to very high concentrations in normal human fibroblasts infected by adenovirus type 5 mutants that cannot direct assembly of the viral E1B 55-kDa protein-containing E3 ubiquitin ligase that targets p53 for degradation. Despite high concentrations of nuclear p53, the p53 transcriptional program is not induced in these infected cells. We exploited this system to examine select post-translational modifications (PTMs) present on a transcriptionally inert population of endogenous human p53, as well as on p53 activated in response to etoposide treatment of normal human fibroblasts. These forms of p53 were purified from whole cell lysates by means of immunoaffinity chromatography and SDS-PAGE, and peptides derived from them were subjected to nano-ultra-high-performance LC-MS and MS/MS analyses on a high-resolution accurate-mass MS platform (data available via ProteomeXchange, PXD000464). We identified an unexpectedly large number of PTMs, comprising phosphorylation of Ser and Thr residues, methylation of Arg residues, and acetylation, ubiquitinylation, and methylation of Lys residues-for example, some 150 previously undescribed modifications of p53 isolated from infected cells. These modifications were distributed across all functional domains of both forms of the endogenous human p53 protein, as well as those of an orthologous population of p53 isolated from COS-1 cells. Despite the differences in activity, including greater in vitro sequence-specific DNA binding activity exhibited by p53 isolated from etoposide-treated cells, few differences were observed in the location, nature, or relative frequencies of PTMs on the two populations of human p53. Indeed, the wealth of PTMs that we have identified is consistent with a far greater degree of complex, combinatorial regulation of p53 by PTM than previously anticipated.
Figures
Fig. 1.
Properties of p53 isolated from normal human fibroblasts. A, HFFs were infected with 100 pfu/cell AdEasy E1 (WT) or AdEasyE1Δ2347 (ΔE1B) for the periods indicated, or mock infected (M). Whole cell extracts were prepared and the proteins listed were examined via immunoblotting as described under “Experimental Procedures.” B, p53 present in HFFs infected with AdEasyE1Δ2347 for 44 h was examined using the antibodies indicated at the top. C, Whole cell extracts were prepared from cells infected with AdEasyE1Δ2347 (ΔE1B) or exposed to 125 μ
m
etoposide (E) for 44 h, and the quantity of p53 present in the increasing extract volumes indicated at the top was compared via immunoblotting.
Fig. 2.
Analysis of DNA binding by p53. The relative concentrations of p53 present in whole cell extracts of HFFs infected with 100 pfu/cell AdEasyE1(Δ)2347 (ΔE1B p53) or exposed to 125 μ
m
etoposide (E p53) for 44 h were determined via immunoblotting and quantification of signals, with β-actin as an internal control. The binding of equal quantities of the two forms of p53 to a consensus DNA binding site was then examined as a function of p53 concentration, as described under “Experimental Procedures.” All panels show the means and standard deviations of triplicate technical replicates. The results of independent experimental replicates are shown in panels A and C, and the initial portion of the binding curve shown in panel A is expanded in panel B.
Fig. 3.
Examples of immunoaffinity purification of p53. HFFs were infected with 100 p.f.u./cell AdEasyE1Δ2347 for 44 h, and following lysate preparation, p53 was purified via immunoaffinity chromatography. A, lysates were prepared in buffer containing 1% (v/v) Triton X-100, and p53 was eluted in 200 m
m
TEA. The relative concentrations of p53, β-actin, and IgG heavy chain (IgG) in the lysate (L), flow-through (F), wash fractions (W1–W3), and eluates (E1 and E2) were determined via immunoblotting. B, lysates were prepared in buffer containing 1% (v/v) N-laurylsarcosine, and p53 was eluted from the immunoaffinity matrix via heat treatment. p53 and IgG heavy chain were visualized as described in panel A.
Fig. 4.
Representative MS/MS spectra of ΔE1B p53. Peptides prepared from p53 isolates were subjected to reversed-phase nano-LC-MS and MS/MS on a UPLC-Orbitrap Velos platform as described under “Experimental Procedures.” Shown are representative examples of tandem mass spectra displaying their PTM-bearing peptide fragment ion assignments. Prominent fragment ions are labeled with their empirical m/z values and b- and y-ion designations or annotations indicating neutral losses from precursor species. The peptide sequences are displayed above the spectra, with all of the fragment ions matched in the spectra indicated by flags. Modified residues are color-coded by PTM in the following manner: phosphorylation, red; acetylation, green; ubiquitinylation, purple; monomethylation, navy; dimethylation, cerulean; trimethylation, turquoise; oxidation, tan; and carbamidomethylation, orange.
Fig. 5.
Representative MS/MS spectra of Ε p53. Peptides prepared from p53 isolated from HFFs treated with etoposide were subjected to reversed-phase nano-LC-MS and MS/MS on a UPLC-Orbitrap Velos platform as described under “Experimental Procedures.” Shown are representative examples of tandem mass spectra displaying their PTM-bearing peptide fragment ion assignments, labeled as described in the legend to Fig. 4.
Fig. 6.
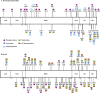
Post-translational modification of p53 isolated from AdEasyE1Δ2347-infected HFFs. The modifications reproducibly identified across three independent biological replicate samples of ΔE1B p53 are summarized, classified according to whether they have been previously described (A) or are novel (B). AD, activation domain; DBD, DNA-binding domain; NLS, nuclear localization signal; TD, tetramerization domain; BD, basic domain.
Fig. 7.
Representative MS/MS spectra of COS-1 p53. Peptides prepared from p53 isolated from COS-1 cells were subjected to reversed-phase nano-LC-MS and MS/MS on a UPLC-Orbitrap Velos platform as described under “Experimental Procedures.” Shown are representative examples of tandem mass spectra displaying their PTM-bearing peptide fragment ion assignments, labeled as described in the legend to Fig. 4.
Fig. 8.
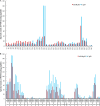
Spectral count profiles of modified Ser, Thr, and Arg residues. Peptides from equivalent populations of ΔE1B p53 and E p53 isolated in parallel were subjected to nano-UPLC-MS and MS/MS analyses on the Orbitrap Velos platform. Modification profiles were generated from technical triplicate LC-MS runs for each sample by means of spectral counting–based quantification of matched PTMs per residue, as described under “Experimental Procedures.” Shown are the modification profiles for (A) phosphorylation of Ser and Thr residues and (B) mono-, di-, and trimethylation of Arg residues present on both p53 populations.
Similar articles
- Impact of the adenoviral E4 Orf3 protein on the activity and posttranslational modification of p53.
DeHart CJ, Perlman DH, Flint SJ. DeHart CJ, et al. J Virol. 2015 Mar;89(6):3209-20. doi: 10.1128/JVI.03072-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568206 Free PMC article. - Identification of new p53 acetylation sites in COS-1 cells.
Joubel A, Chalkley RJ, Medzihradszky KF, Hondermarck H, Burlingame AL. Joubel A, et al. Mol Cell Proteomics. 2009 Jun;8(6):1167-73. doi: 10.1074/mcp.M800487-MCP200. Epub 2009 Jan 19. Mol Cell Proteomics. 2009. PMID: 19155208 Free PMC article. - E1B-55K-Mediated Regulation of RNF4 SUMO-Targeted Ubiquitin Ligase Promotes Human Adenovirus Gene Expression.
Müncheberg S, Hay RT, Ip WH, Meyer T, Weiß C, Brenke J, Masser S, Hadian K, Dobner T, Schreiner S. Müncheberg S, et al. J Virol. 2018 Jun 13;92(13):e00164-18. doi: 10.1128/JVI.00164-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29695423 Free PMC article. - p53 modifications: exquisite decorations of the powerful guardian.
Liu Y, Tavana O, Gu W. Liu Y, et al. J Mol Cell Biol. 2019 Jul 19;11(7):564-577. doi: 10.1093/jmcb/mjz060. J Mol Cell Biol. 2019. PMID: 31282934 Free PMC article. Review. - [The role of different E3 ubiquitin ligases in regulation of the P53 tumor suppressor protein].
Daks AA, Melino D, Barlev NA. Daks AA, et al. Tsitologiia. 2013;55(10):673-87. Tsitologiia. 2013. PMID: 25509121 Review. Russian.
Cited by
- Structural plasticity of methyllysine recognition by the tandem tudor domain of 53BP1.
Tong Q, Cui G, Botuyan MV, Rothbart SB, Hayashi R, Musselman CA, Singh N, Appella E, Strahl BD, Mer G, Kutateladze TG. Tong Q, et al. Structure. 2015 Feb 3;23(2):312-21. doi: 10.1016/j.str.2014.11.013. Epub 2015 Jan 8. Structure. 2015. PMID: 25579814 Free PMC article. - Shaping the regulation of the p53 mRNA tumour suppressor: the co-evolution of genetic signatures.
Karakostis K, Fåhraeus R. Karakostis K, et al. BMC Cancer. 2019 Sep 13;19(1):915. doi: 10.1186/s12885-019-6118-y. BMC Cancer. 2019. PMID: 31519161 Free PMC article. Review. - Posttranslational Protein Modifications in Plant Metabolism.
Friso G, van Wijk KJ. Friso G, et al. Plant Physiol. 2015 Nov;169(3):1469-87. doi: 10.1104/pp.15.01378. Epub 2015 Sep 3. Plant Physiol. 2015. PMID: 26338952 Free PMC article. - Lysines in the tetramerization domain of p53 selectively modulate G1 arrest.
Beckerman R, Yoh K, Mattia-Sansobrino M, Zupnick A, Laptenko O, Karni-Schmidt O, Ahn J, Byeon IJ, Keezer S, Prives C. Beckerman R, et al. Cell Cycle. 2016 Jun 2;15(11):1425-38. doi: 10.1080/15384101.2016.1170270. Epub 2016 May 21. Cell Cycle. 2016. PMID: 27210019 Free PMC article. - Knockdown of TFAM in Tumor Cells Retarded Autophagic Flux through Regulating p53 Acetylation and PISD Expression.
Jiang X, Wang J. Jiang X, et al. Cancers (Basel). 2020 Feb 20;12(2):493. doi: 10.3390/cancers12020493. Cancers (Basel). 2020. PMID: 32093281 Free PMC article.
References
- Lane D. P., Crawford L. V. (1979) T antigen is bound to a host protein in SY40-transformed cells. Nature 278, 261–263 - PubMed
- Linzer D. I., Levine A. J. (1979) Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17, 43–52 - PubMed
- Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 - PubMed
- Harris S. L., Levine A. J. (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous