Arabidopsis clade I TGA factors regulate apoplastic defences against the bacterial pathogen Pseudomonas syringae through endoplasmic reticulum-based processes - PubMed (original) (raw)
Arabidopsis clade I TGA factors regulate apoplastic defences against the bacterial pathogen Pseudomonas syringae through endoplasmic reticulum-based processes
Lipu Wang et al. PLoS One. 2013.
Abstract
During the plant immune response, large-scale transcriptional reprogramming is modulated by numerous transcription (co) factors. The Arabidopsis basic leucine zipper transcription factors TGA1 and TGA4, which comprise the clade I TGA factors, have been shown to positively contribute to disease resistance against virulent strains of the bacterial pathogen Pseudomonas syringae. Despite physically interacting with the key immune regulator, NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1), following elicitation with salicylic acid (SA), clade I function was shown to be largely independent of NPR1. Unlike mutants in NPR1, tga1-1 tga4-1 plants do not display reductions in steady-state levels of SA-pathway marker genes following treatment with this phenolic signaling metabolite or after challenge with virulent or avirulent P. syringae. By exploiting bacterial strains that have limited capacity to suppress Arabidopsis defence responses, the present study demonstrates that tga1-1 tga4-1 plants are compromised in basal resistance and defective in several apoplastic defence responses, including the oxidative burst of reactive oxygen species, callose deposition, as well as total and apoplastic PATHOGENESIS-RELATED 1 (PR-1) protein accumulation. Furthermore, analysis of npr1-1 and the tga1-1 tga4-1 npr1-1 triple mutant indicates that clade I TGA factors act substantially independent of NPR1 in mediating disease resistance against these strains of P. syringae. Increased sensitivity to the N-glycosylation inhibitor tunicamycin and elevated levels of endoplasmic reticulum (ER) stress marker genes encoding ER-resident chaperones in mutant seedlings suggest that loss of apoplastic defence responses is associated with aberrant protein secretion and implicate clade I TGA factors as positive regulators of one or more ER-related secretion pathways.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
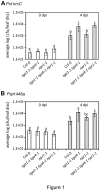
Figure 1. Growth of Pst hrcC - (A) and Psp 1448a (B) in Col-0, tga1-1 tga4-1, npr1-1 and tga1-1 tga4-1 npr1-1 plants.
Four-week old leaves were syringe-infiltrated with a bacterial suspension of Pst hrcC - at 105 colony forming units (cfu) ml-1 or Psp 1448a at 106 cfu ml-1. The error bars represent the standard deviation of six replicates, each containing 8 leaf discs from one plant. An ANOVA of the log-transformed data was performed at α = 0.05; treatments with common letters over bars are not significantly different from each other. Post-hoc tests are presented in Table S2. Each experiment was repeated twice with similar results.
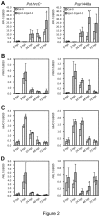
Figure 2. Defence-related gene expression in Col-0 and tga1-1 tga4-1.
Four-week-old leaves were syringe-infiltrated with 108 cfu ml-1 of Pst hrcC - or Psp 1448a. Leaf tissues from three plants were collected and pooled as one sample for RNA isolation. Values were normalized to the expression of UBIQUITIN5. An asterisk indicates a statistically significant difference compared with Col-0 at the same time point (p<0.05, Student’s _t_-test), and two asterisks indicate p<0.01. The error bars represent the standard deviation of three biological samples.
Figure 3. MAMPs- and pathogen-induced callose deposition in Col-0 and tga1-1 tga4-1 plants.
Four-week-old leaves were syringe-infiltrated with 108 cfu ml-1 of Pst hrcC - or Psp 1448a, 5µM flg22, and 10mM MgCl2 as control. Leaves were stained with Aniline blue and observed under a florescent microscope 12 h after treatment. Microscopic photographs of callose deposits are shown with the number of callose deposits indicated below each. Results are presented as means ± standard deviation. An asterisk indicates a statistically significant difference between Col-0 and tga1-1 tga4-1 (p<0.05, Student’s _t_-test), and two asterisks indicate p<0.01. Each experiment was repeated three times with similar results. Scale bar = 0.1mm, all photos are at the same magnification.
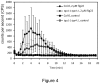
Figure 4. MAMPs-induced oxidative burst in Col-0 and tga1-1tga4-1 plants.
Four-week-old leaf discs (3 per each sample) were treated with or without 2 µM flg22 in the presence of luminol, and the H2O2 generated was measured every 30 sec after treatment for 20 min. The error bars represent the standard deviation of six replicates. The experiment was repeated five times with similar results.
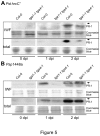
Figure 5. PR-1 protein accumulation in Col-0 and tga1-1 tga4-1 after pathogen inoculation.
Four-week-old leaves were syringe-infiltrated with 108 cfu ml-1 of Pst hrcC - or Psp 1448a. Intercellular washing fluids (IWFs) and total protein were collected at 0, 1 and 2 dpi, separated on 16% Tricine-SDS-polyacrylamide gels and blotted with a PR-1 antibody. The Arabidopsis PR-1 protein has apredicted molecular weight of 16 kilodaltons (kDa) [74]. Arrows indicate the position of a 17-kDa molecular weight masker. The same gels were stained with Coomassie Brilliant Blue R250 (Sigma) as a loading control. These experiments were repeated three times with similar results.
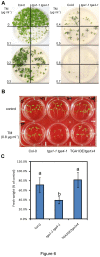
Figure 6. Tunicamycin sensitivity of Col-0 and the tga1-1 tga4-1 plants.
A, Five-day-old seedlings of Col-0 and the tga1-1 tga4-1 double mutant grown on Murashige and Skoog (MS) agar medium with different concentrations of TM were transplanted to TM-free MS agar and grown for a further 5 days prior to photography. This experiment was repeated three times with similar results. B, Five-day-old seedlings of Col-0, tga1-1 tga4-1, and a line overexpressing TGA1 in the tga1-1 tga4-1 background (TGA1OE/tga1x4) grown on TM-free MS agar were submerged in MS liquid medium with or without 0.8 µg ml-1 TM for 6 h, and were allowed to recover for 5 days without TM prior to photography. C, Fresh weight of seedlings in (B) was quantified. The fresh weight of TM-treated seedlings was divided by the average fresh weight of 5 untreated seedlings to generate percentage of control. The results are averages ± standard deviation (n=5). An ANOVA of data was performed at α = 0.05; treatments with common letters over the error bars are not significantly different from each other. This experiment was repeated twice with similar results.
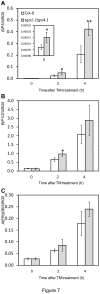
Figure 7. ER stress gene expression in Col-0 and the tga1-1 tga4-1 plants.
Ten-day-old seedlings were immersed with 5 µg ml-1 TM for the indicated time periods. Fifty mg samples were collected and pooled for RNA isolation. Values were normalized to the expression of UBIQUITIN5. An asterisk indicates a statistically significant difference compared with Col-0 at the same time point (p<0.05, Student’s _t_-test), and two asterisks indicate p<0.01. The error bars represent the standard deviation of four biological samples. The oligonucleotides used in (B) did not distinguish between BiP1 and BiP2. Accordlingly, the target is referred to as BiP1/2.
Similar articles
- Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion.
Shearer HL, Cheng YT, Wang L, Liu J, Boyle P, Després C, Zhang Y, Li X, Fobert PR. Shearer HL, et al. Mol Plant Microbe Interact. 2012 Nov;25(11):1459-68. doi: 10.1094/MPMI-09-11-0256. Mol Plant Microbe Interact. 2012. PMID: 22876961 - Redox-active cysteines in TGACG-BINDING FACTOR 1 (TGA1) do not play a role in salicylic acid or pathogen-induced expression of TGA1-regulated target genes in Arabidopsis thaliana.
Budimir J, Treffon K, Nair A, Thurow C, Gatz C. Budimir J, et al. New Phytol. 2021 Jun;230(6):2420-2432. doi: 10.1111/nph.16614. Epub 2020 Jun 16. New Phytol. 2021. PMID: 32315441 - NPR1: the spider in the web of induced resistance signaling pathways.
Pieterse CM, Van Loon LC. Pieterse CM, et al. Curr Opin Plant Biol. 2004 Aug;7(4):456-64. doi: 10.1016/j.pbi.2004.05.006. Curr Opin Plant Biol. 2004. PMID: 15231270 Review. - Singlet oxygen-mediated and EXECUTER-dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress.
Zhang S, Apel K, Kim C. Zhang S, et al. Philos Trans R Soc Lond B Biol Sci. 2014 Mar 3;369(1640):20130227. doi: 10.1098/rstb.2013.0227. Print 2014 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 24591714 Free PMC article. Review.
Cited by
- Divergence of functions and expression patterns of soybean bZIP transcription factors.
Yue L, Pei X, Kong F, Zhao L, Lin X. Yue L, et al. Front Plant Sci. 2023 Apr 14;14:1150363. doi: 10.3389/fpls.2023.1150363. eCollection 2023. Front Plant Sci. 2023. PMID: 37123868 Free PMC article. Review. - The Potyviral P3 Protein Targets Eukaryotic Elongation Factor 1A to Promote the Unfolded Protein Response and Viral Pathogenesis.
Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A. Luan H, et al. Plant Physiol. 2016 Sep;172(1):221-34. doi: 10.1104/pp.16.00505. Epub 2016 Jun 29. Plant Physiol. 2016. PMID: 27356973 Free PMC article. - Synchronization of developmental processes and defense signaling by growth regulating transcription factors.
Liu J, Rice JH, Chen N, Baum TJ, Hewezi T. Liu J, et al. PLoS One. 2014 May 29;9(5):e98477. doi: 10.1371/journal.pone.0098477. eCollection 2014. PLoS One. 2014. PMID: 24875638 Free PMC article. - Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming.
Seyfferth C, Tsuda K. Seyfferth C, et al. Front Plant Sci. 2014 Dec 9;5:697. doi: 10.3389/fpls.2014.00697. eCollection 2014. Front Plant Sci. 2014. PMID: 25538725 Free PMC article. - Insight into the bZIP gene family in Lagenaria siceraria: Genome and transcriptome analysis to understand gene diversification in Cucurbitaceae and the roles of LsbZIP gene expression and function under cold stress.
Wang J, Wang Y, Wu X, Wang B, Lu Z, Zhong L, Li G, Wu X. Wang J, et al. Front Plant Sci. 2023 Feb 17;13:1128007. doi: 10.3389/fpls.2022.1128007. eCollection 2022. Front Plant Sci. 2023. PMID: 36874919 Free PMC article.
References
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323-329. doi:10.1038/nature05286. PubMed: 17108957. - DOI - PubMed
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379-406. doi:10.1146/annurev.arplant.57.032905.105346. PubMed: 19400727. - DOI - PubMed
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742-744. doi:10.1126/science.1171647. PubMed: 19423812. - DOI - PMC - PubMed
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185-209. doi:10.1146/annurev.phyto.42.040803.140421. PubMed: 15283665. - DOI - PubMed
- Dempsey DA, Klessig DF (2012) SOS – too many signals for systemic acquired resistance? Trends Plant Sci 17: 538-545. doi:10.1016/j.tplants.2012.05.011. PubMed: 22749315. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous