RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia - PubMed (original) (raw)
Clinical Trial
. 2013 Dec 17;110(51):E4968-77.
doi: 10.1073/pnas.1315438110. Epub 2013 Nov 18.
Yuanjing Liu, Monica Bañez-Coronel, Tammy Reid, Olga Pletnikova, Jada Lewis, Timothy M Miller, Matthew B Harms, Annet E Falchook, S H Subramony, Lyle W Ostrow, Jeffrey D Rothstein, Juan C Troncoso, Laura P W Ranum
Affiliations
- PMID: 24248382
- PMCID: PMC3870665
- DOI: 10.1073/pnas.1315438110
Clinical Trial
RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia
Tao Zu et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The finding that a GGGGCC (G4C2) hexanucleotide repeat expansion in the chromosome 9 ORF 72 (C9ORF72) gene is a common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) links ALS/FTD to a large group of unstable microsatellite diseases. Previously, we showed that microsatellite expansion mutations can be bidirectionally transcribed and that these mutations express unexpected proteins by a unique mechanism, repeat-associated non-ATG (RAN) translation. In this study, we show that C9ORF72 antisense transcripts are elevated in the brains of C9ORF72 expansion-positive [C9(+)] patients, and antisense GGCCCC (G2C4) repeat-expansion RNAs accumulate in nuclear foci in brain. Additionally, sense and antisense foci accumulate in blood and are potential biomarkers of the disease. Furthermore, we show that RAN translation occurs from both sense and antisense expansion transcripts, resulting in the expression of six RAN proteins (antisense: Pro-Arg, Pro-Ala, Gly-Pro; and sense: Gly-Ala, Gly-Arg, Gly-Pro). These proteins accumulate in cytoplasmic aggregates in affected brain regions, including the frontal and motor cortex, hippocampus, and spinal cord neurons, with some brain regions showing dramatic RAN protein accumulation and clustering. The finding that unique antisense G2C4 RNA foci and three unique antisense RAN proteins accumulate in patient tissues indicates that bidirectional transcription of expanded alleles is a fundamental pathologic feature of C9ORF72 ALS/FTD. Additionally, these findings suggest the need to test therapeutic strategies that target both sense and antisense RNAs and RAN proteins in C9ORF72 ALS/FTD, and to more broadly consider the role of antisense expression and RAN translation across microsatellite expansion diseases.
Keywords: clustered aggregates; cytoplasmic inclusions; noncoding RNA.
Conflict of interest statement
Conflict of interest statement: T.Z. and L.P.W.R. are listed as inventors on pending patents on RAN proteins.
Figures
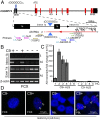
Fig. 1.
G2C4 antisense transcripts accumulate as RNA foci in C9ORF72 patient tissues. (A) Schematic diagram of C9ORF72 gene and antisense transcripts and relative location of primers for strand-specific RT-PCR and RACE primers. (B) Strand-specific RT-PCR of sense (S) and antisense (AS) transcripts (across intron 1b and exon 1a) from FCX of C9(+) and C9(−) ALS patients. (C) Strand-specific qRT-PCR showing elevated antisense mRNA in C9(+) compared with C9(−) ALS patients and a healthy control (HC). (D) FISH with G4C2-Cy3 probe showing G2C4 antisense RNA foci (red) in C9(+)FCX and PBLs which are absent in C9(−) cases. Nuclear foci in FCX are indicated by arrowheads.
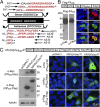
Fig. 2.
In vitro evidence for RAN translation of antisense G2C4 expansion and dual immunological detection strategy. (A) Diagram of putative proteins translated from sense and antisense transcripts. Sequences highlighted in red were used to generate antibodies. CT, C-terminal; f1–3, reading frames 1–3; *, stop codon. (B) Abbreviated example of validation of α-PAAS rabbit polyclonal antibody. Immunoblots (Lower Left) and IF staining (Lower Right) of HEK293T cells transfected with Flag-PAAS construct (Upper) and probed with α-Flag and α-PAAS antibodies. See
Figs. S4
and
S5
for additional controls and validation of eight additional antibodies generated against RAN repeat motifs and CT regions. (C) Immunoblots (Lower Left) and IF staining (Lower Right) of HEK293T cells 48 h posttransfection with the AS-(G2C4)EXP-3T construct (Upper). PRAS and GPAS expansion proteins were detected by Western blot and PAAS, PRAS and GPAS proteins were detected by IF in transfected cells.
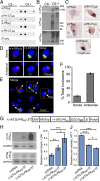
Fig. 3.
In vivo evidence for RAN-translation of the G2C4 AS repeat and toxicity studies. (A) Dot blots containing frontal cortex lysates from four different C9(−) and four different C9(+) ALS/FTD patients were probed with α-PAAS, α-PA-CTAS, α-PRAS, and α-PR-CTAS antibodies. Results show evidence of RAN protein accumulation with all four antibodies in two of four C9(+) samples and weaker positive staining in one sample with α-PAAS. (B) Immunoblots of C9(+) and C9(−) ALS frontal cortex 2% SDS insoluble fractions with α-PRAS, α-PAAS, and α-GPS/AS. (C) IHC detection of PAAS, PRAS, and GPS/AS protein aggregates in hippocampal neurons from C9(+) ALS patients detected with α-PAAS, α-PA-CTAS, α-PRAS, α-PR-CTAS, and α-GPS/AS antibodies. (D) IF staining with mouse α-GPS/AS (Cy3, red) and rabbit α-GP-CTS (Alexa Fluor 488, green) of C9(+)hippocampal tissue with sense inclusions positive for both antibodies (Upper) and antisense inclusions positive for only GP repeat antibody (Lower). Colocalization of α-GPS/AS and α-GP-CTS staining was confirmed by confocal _z_-stacks. (E) IF staining of larger region. Arrowheads indicate background lipofuscin autofluroescense, which can be distinguished from the real signal because it fluoresces in all three channels and when merged appears as brownish-orange signal that is distinct from the Cy3 and Alexa Fluor 488 signals shown (31). (F) Percentages of double- (sense) and single- (antisense) labeled aggregates were quantitated among 1,389 cells from three different tiled areas and a total of 88 aggregates. (G–J) RAN and PRAS toxicity studies. (G) G2C4 expansion constructs (+/−ATG-PR-3T) with or without an ATG initiation codon in PRAS frame and 3′ epitope tags. (H) Western blots showing levels of PRAS and GPAS in cells transfected with constructs in G. (I) LDH and (J) MTT assays of transfected HEK293T cells. *P ≤ 0.05, **P ≤ 0.01), ***P ≤ 0.001, n = 6 independent experiments).
Fig. 4.
In vivo evidence for RAN translation in both antisense and sense directions of C9ORF72. Cytoplasmic inclusions detected by IHC using antibodies against sense (α-GRS, α-GR-CTS, α-GA-CTS, α-GP-CTS) and antisense (α-PAAS, α-PA-CTAS, α-PRAS, α-PR-CTAS), and α-GPS/AS, which recognizes GP proteins made in both the sense and antisense directions. Aggregates are found in neurons of the CA and DG regions of the hippocampus and the motor cortex (MC) of C9(+) ALS autopsy tissue.
Fig. 5.
Clustered RAN-positive cells in hippocampus and motor cortex and RAN aggregates in motor neurons. IHC showing cytoplasmic α-GPS/AS aggregates in: (A) layer III of motor cortex; (B) upper motor neuron in layer V of the motor cortex; (C) lower motor neurons in the lumbar spinal cord (LSC); (D) in CA and (E) DG regions of the hippocampus. (F and G) Abundant PA-CTAS and PR-CTAS cytoplasmic inclusions in the presubiculum (PrSub) from one patient.
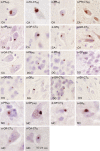
Fig. 6.
Clustered staining of RAN proteins. (A) Low-power image of IHC staining with α-PA-CTAS shows variations in staining intensity (red is positive) in regions I–IV, with Insets showing higher-power images. (B) Examples of aggregates from region I show immunoreactivity against all nine antibodies with similar staining for antibodies against repeat and unique C-terminal epitopes.
Similar articles
- Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS.
Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Gendron TF, et al. Acta Neuropathol. 2013 Dec;126(6):829-44. doi: 10.1007/s00401-013-1192-8. Epub 2013 Oct 16. Acta Neuropathol. 2013. PMID: 24129584 Free PMC article. - Antibody Therapy Targeting RAN Proteins Rescues C9 ALS/FTD Phenotypes in C9orf72 Mouse Model.
Nguyen L, Montrasio F, Pattamatta A, Tusi SK, Bardhi O, Meyer KD, Hayes L, Nakamura K, Banez-Coronel M, Coyne A, Guo S, Laboissonniere LA, Gu Y, Narayanan S, Smith B, Nitsch RM, Kankel MW, Rushe M, Rothstein J, Zu T, Grimm J, Ranum LPW. Nguyen L, et al. Neuron. 2020 Feb 19;105(4):645-662.e11. doi: 10.1016/j.neuron.2019.11.007. Epub 2019 Dec 9. Neuron. 2020. PMID: 31831332 Free PMC article. - Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.
Wen X, Westergard T, Pasinelli P, Trotti D. Wen X, et al. Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review. - RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention.
Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. Donnelly CJ, et al. Neuron. 2013 Oct 16;80(2):415-28. doi: 10.1016/j.neuron.2013.10.015. Neuron. 2013. PMID: 24139042 Free PMC article. - Disease Mechanisms of C9ORF72 Repeat Expansions.
Gendron TF, Petrucelli L. Gendron TF, et al. Cold Spring Harb Perspect Med. 2018 Apr 2;8(4):a024224. doi: 10.1101/cshperspect.a024224. Cold Spring Harb Perspect Med. 2018. PMID: 28130314 Free PMC article. Review.
Cited by
- SETX (senataxin), the helicase mutated in AOA2 and ALS4, functions in autophagy regulation.
Richard P, Feng S, Tsai YL, Li W, Rinchetti P, Muhith U, Irizarry-Cole J, Stolz K, Sanz LA, Hartono S, Hoque M, Tadesse S, Seitz H, Lotti F, Hirano M, Chédin F, Tian B, Manley JL. Richard P, et al. Autophagy. 2021 Aug;17(8):1889-1906. doi: 10.1080/15548627.2020.1796292. Epub 2020 Aug 7. Autophagy. 2021. PMID: 32686621 Free PMC article. - Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72-ALS/FTD.
Westergard T, Jensen BK, Wen X, Cai J, Kropf E, Iacovitti L, Pasinelli P, Trotti D. Westergard T, et al. Cell Rep. 2016 Oct 11;17(3):645-652. doi: 10.1016/j.celrep.2016.09.032. Cell Rep. 2016. PMID: 27732842 Free PMC article. - Staufen Impairs Autophagy in Neurodegeneration.
Paul S, Dansithong W, Gandelman M, Figueroa KP, Zu T, Ranum LPW, Scoles DR, Pulst SM. Paul S, et al. Ann Neurol. 2023 Feb;93(2):398-416. doi: 10.1002/ana.26515. Epub 2022 Oct 27. Ann Neurol. 2023. PMID: 36151701 Free PMC article. - The conserved multi-functional nuclear protein dss-1/Sem1 is required for C9orf72-associated ALS/FTD dipeptide toxicity.
Puleo N, Lamitina T. Puleo N, et al. MicroPubl Biol. 2020 Jun 2;2020:10.17912/micropub.biology.000262. doi: 10.17912/micropub.biology.000262. MicroPubl Biol. 2020. PMID: 32550521 Free PMC article. No abstract available. - Multifaceted Genes in Amyotrophic Lateral Sclerosis-Frontotemporal Dementia.
Ranganathan R, Haque S, Coley K, Shepheard S, Cooper-Knock J, Kirby J. Ranganathan R, et al. Front Neurosci. 2020 Jul 7;14:684. doi: 10.3389/fnins.2020.00684. eCollection 2020. Front Neurosci. 2020. PMID: 32733193 Free PMC article. Review.
References
- Majounie E, et al. Chromosome 9-ALS/FTD Consortium French Research Network on FTLD/FTLD/ALS ITALSGEN Consortium Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012;11(4):323–330. - PMC - PubMed
- Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012;11(1):54–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS085207/NS/NINDS NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- K08 NS075094/NS/NINDS NIH HHS/United States
- R01 NS040389/NS/NINDS NIH HHS/United States
- NIH P50AG05146/AG/NIA NIH HHS/United States
- P01 NS058901/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous