Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase - PubMed (original) (raw)
Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase
Young-Hoon Kang et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Chromosome transmission fidelity 4 (Ctf4) is a conserved protein required for DNA replication. In this report, interactions between human Ctf4 (hCtf4) and the replicative helicase containing the cell division cycle 45 (Cdc45)/minichromosome maintenance 2-7 (Mcm2-7)/Go, Ichi, Nii, and San (GINS) (CMG) proteins [human CMG (hCMG) complex] were examined. The hCtf4-CMG complex was isolated following in vitro interaction of purified proteins (hCtf4 plus the hCMG complex), coinfection of Spodoptera frugiperda (Sf9) insect cells with viruses expressing the hCMG complex and hCtf4, and from HeLa cell chromatin after benzonase and immunoprecipitation steps. The stability of the hCtf4-CMG complex depends upon interactions between hCtf4 and multiple components of the hCMG complex. The hCtf4-CMG complex, like the hCMG complex, contains DNA helicase activity that is more salt-resistant than the helicase activity of the hCMG complex. We demonstrate that the hCtf4-CMG complex contains a homodimeric hCtf4 and a monomeric hCMG complex and suggest that the homodimeric hCtf4 acts as a platform linking polymerase α to the hCMG complex. The role of the hCMG complex as the core of the replisome is also discussed.
Keywords: dimerization; replication initiation; replisome core structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
hCtf4 forms a stable complex with the hCMG complex. (A) Purified hCMG (5 pmol) (HF tag on Cdc45) and untagged hCtf4 (15 pmol, as a dimer) were mixed with anti-FLAG M2 affinity gel. (Upper) Bound proteins were eluted and subjected to glycerol gradient sedimentation. (Lower) As a control, untagged hCtf4 was sedimented separately. CBB, Coomassie brilliant blue. The hCtf4–CMG complex was purified as described in
SI Materials and Methods
, and each fraction (5 μL) was separated electrophoretically through a 4–20% (wt/vol) polyacrylamide gradient gel (Invitrogen), followed by silver staining (B) or Western blot analysis (C) against all 12 subunits of the hCtf4–CMG complex. The peak positions of protein markers (GE Healthcare Life Sciences) after sedimentation are indicated above fractions (669 kDa, thyroglobulin; 158 kDa, aldolase; 43 kDa, ovalbumin). (D) hCtf4–CMG and hCMG were loaded side by side onto a 4–20% (wt/vol) gel and silver-stained. (E) Helicase activity was measured across the hCtf4–CMG glycerol gradient (0.5-μL fractions), and the substrate unwound (%) was calculated and is shown in the graph. The structure of the helicase substrate containing a 57-mer duplex region and a 5′-dT40 tail (M13 annealed to labeled oligonucleotide no. 2) is shown in the graph. See
Table S1
for description of oligonucleotides in this study. *, Location of 32P in substrate. B, boiled substrate.
Fig. 2.
hCtf4 stimulates the hCMG helicase activity. Indicated levels of hCMG and untagged hCtf4 were incubated using helicase assay conditions containing 5 mM NaCl (A) or 50 mM NaCl (B). (C) Helicase assays were performed with the hCMG and hCtf4–CMG complexes in the presence of 5 or 50 mM NaCl. (D) Substrate unwound (%) from the results presented in C was plotted against the protein added (femtomoles). It should be noted that the amount of hCtf4 indicated in the figures was calculated as a dimer.
Fig. 3.
hCtf4 is present as a dimer in the hCtf4–CMG complex. Sf9 cells were infected with two differently tagged hCtf4 viruses (H-hCtf4 and F-hCtf4) and viruses expressing hCMG components, and the free hCtf4 and hCtf4–CMG complex formed were purified. Western blot analyses of fractions from the glycerol gradient sedimentation of hCtf4 (A) and hCtf4–CMG (B) are shown. The peak glycerol gradient fractions of the hCtf4 and hCtf4–CMG preparations, shown in A and B, were loaded onto a 4–20% polyacrylamide gel side by side with hCtf4 (C) or hCtf4–CMG (D) purified from cells expressing F-hCtf4 and the hCMG complex. Regions containing hCtf4 bands were cropped in these figures (images from the original gels are shown in
Fig. S4 E and F
). (E) Sf9 cells were coinfected with F-hCtf4 and hCMG viruses that included two differently tagged Sld5 subunits (GST-Sld5 and H-Sld5), and the hCtf4–CMG complex formed was purified. The peak glycerol gradient fraction was loaded onto gels side by side with untagged and H-hGINS (His6 tag on Sld5) and immunoblotted for Sld5.
Fig. 4.
Physical and biochemical interactions between different domains of hCtf4 and the hCMG complex. (A) Various derivatives of HF-hCtf4 [300 fmol of full-length, N (WD + SepB), C (SepB + HMG), and SepB domains, which are likely to be dimers, and 600 fmol of WD and HMG domain, which are likely to be monomers] were incubated in the presence (+) or absence (−) of the hCMG complex (100 fmol). Mixtures were then immunoprecipitated with 1 μg of α-Cdc45 antibodies. Loading controls and immunoprecipitated materials were gel-separated and then analyzed by Western blotting against the FLAG tag to detect HF-hCtf4 derivatives and HF-hCdc45. The domains of hCtf4 are shown below the gel. (B) HF-hCtf4 derivatives (50 fmol as dimers and 100 fmol of monomers) used in A and the hCMG complex (100 fmol) were mixed and incubated in standard helicase reaction mixtures containing 50 mM NaCl. Untagged, full-length hCtf4 was also included as a control (last lane). (C) Unwound substrate (%), presented in B, was calculated and is shown in the graph.
Fig. 5.
Comparison of interactions of hCtf4 with the hCMG complex and hMcm2-7, hGINS, or hCdc45. (A) hCMG or hCdc45 (100 fmol) was incubated with hCtf4 (400 fmol), and the mixture was immunoprecipitated with 1 μg of α-Cdc45. The precipitated material and various levels of input proteins were separated by 10% (wt/vol) SDS/PAGE and immunoblotted against the indicated proteins. (B) hCMG or hMcm2-7 (100 fmol) was incubated with hCtf4 (400 fmol) and immunoprecipitated with 2.5 μg of α-Mcm5. (C) hCMG or hGINS (100 fmol) was incubated with hCtf4 (400 fmol) and immunoprecipitated with 2 μg of α-Sld5. IP, immunoprecipitation.
Fig. 6.
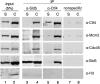
hCMG complex associates with hCtf4 on chromatin in HeLa cells. Soluble (S) or chromatin (C) fractions (500 μg) of protein isolated from HeLa cells were incubated with Sld5 antibodies (lanes 3 and 4), Ctf4 (lanes 5 and 6), or nonspecific (GST) antibodies (lanes 7 and 8), as indicated above the immunoblots. Specific interactions were detected by Western blotting using antibodies to Ctf4, Mcm2, Cdc45, Sld5, or Histone H3. Input represents 5% of the lysate used for immunoprecipitation (lanes 1 and 2).
Similar articles
- TIMELESS Suppresses the Accumulation of Aberrant CDC45·MCM2-7·GINS Replicative Helicase Complexes on Human Chromatin.
Xu X, Wang JT, Li M, Liu Y. Xu X, et al. J Biol Chem. 2016 Oct 21;291(43):22544-22558. doi: 10.1074/jbc.M116.719963. Epub 2016 Sep 1. J Biol Chem. 2016. PMID: 27587400 Free PMC article. - Mcm10 coordinates the timely assembly and activation of the replication fork helicase.
Perez-Arnaiz P, Bruck I, Kaplan DL. Perez-Arnaiz P, et al. Nucleic Acids Res. 2016 Jan 8;44(1):315-29. doi: 10.1093/nar/gkv1260. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26582917 Free PMC article. - A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome.
Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinković D, Labib K, Costa A, Pellegrini L. Simon AC, et al. Nature. 2014 Jun 12;510(7504):293-297. doi: 10.1038/nature13234. Epub 2014 May 4. Nature. 2014. PMID: 24805245 Free PMC article. - The GINS complex: structure and function.
Kamada K. Kamada K. Subcell Biochem. 2012;62:135-56. doi: 10.1007/978-94-007-4572-8_8. Subcell Biochem. 2012. PMID: 22918584 Review. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review.
Cited by
- Replisome function during replicative stress is modulated by histone h3 lysine 56 acetylation through Ctf4.
Luciano P, Dehé PM, Audebert S, Géli V, Corda Y. Luciano P, et al. Genetics. 2015 Apr;199(4):1047-63. doi: 10.1534/genetics.114.173856. Epub 2015 Feb 18. Genetics. 2015. PMID: 25697176 Free PMC article. - The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks.
van Schie JJM, de Lange J. van Schie JJM, et al. Cells. 2021 Dec 8;10(12):3455. doi: 10.3390/cells10123455. Cells. 2021. PMID: 34943967 Free PMC article. Review. - The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy.
Seo YS, Kang YH. Seo YS, et al. Front Mol Biosci. 2018 Mar 29;5:26. doi: 10.3389/fmolb.2018.00026. eCollection 2018. Front Mol Biosci. 2018. PMID: 29651420 Free PMC article. Review. - Rice and Arabidopsis homologs of yeast CHROMOSOME TRANSMISSION FIDELITY PROTEIN 4 commonly interact with Polycomb complexes but exert divergent regulatory functions.
Zhang P, Zhu C, Geng Y, Wang Y, Yang Y, Liu Q, Guo W, Chachar S, Riaz A, Yan S, Yang L, Yi K, Wu C, Gu X. Zhang P, et al. Plant Cell. 2021 Jul 2;33(5):1417-1429. doi: 10.1093/plcell/koab047. Plant Cell. 2021. PMID: 33647940 Free PMC article. - The Involvement of WDHD1 in the Occurrence of Esophageal Cancer as a Downstream Target of PI3K/AKT Pathway.
Xian Q, Zhu D. Xian Q, et al. J Oncol. 2022 Apr 5;2022:5871188. doi: 10.1155/2022/5871188. eCollection 2022. J Oncol. 2022. PMID: 35422862 Free PMC article. Review.
References
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87(1):53–63. - PubMed
- Araki H. Initiation of chromosomal DNA replication in eukaryotic cells; Contribution of yeast genetics to the elucidation. Genes Genet Syst. 2011;86(3):141–149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous