Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell - PubMed (original) (raw)
. 2014 Jan 29;9(1):e86717.
doi: 10.1371/journal.pone.0086717. eCollection 2014.
Behrad Vahidi 2, Norma Arquilevich 1, Cong Fang 1, Samuel Ferguson 2, Darren Harkins 1, Cory Hill 1, Erich Klem 1, Paul C Pagano 1, Chrissy Peasley 1, Juan Romero 1, Robert Shartle 2, Robert C Vasko 1, William M Strauss 1, Paul W Dempsey 1
Affiliations
- PMID: 24489774
- PMCID: PMC3906064
- DOI: 10.1371/journal.pone.0086717
Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell
Jessamine P Winer-Jones et al. PLoS One. 2014.
Abstract
Background: Contemporary cancer diagnostics are becoming increasing reliant upon sophisticated new molecular methods for analyzing genetic information. Limiting the scope of these new technologies is the lack of adequate solid tumor tissue samples. Patients may present with tumors that are not accessible to biopsy or adequate for longitudinal monitoring. One attractive alternate source is cancer cells in the peripheral blood. These rare circulating tumor cells (CTC) require enrichment and isolation before molecular analysis can be performed. Current CTC platforms lack either the throughput or reliability to use in a clinical setting or they provide CTC samples at purities that restrict molecular access by limiting the molecular tools available.
Methodology/principal findings: Recent advances in magetophoresis and microfluidics have been employed to produce an automated platform called LiquidBiopsy®. This platform uses high throughput sheath flow microfluidics for the positive selection of CTC populations. Furthermore the platform quantitatively isolates cells useful for molecular methods such as detection of mutations. CTC recovery was characterized and validated with an accuracy (<20% error) and a precision (CV<25%) down to at least 9 CTC/ml. Using anti-EpCAM antibodies as the capture agent, the platform recovers 78% of MCF7 cells within the linear range. Non specific recovery of background cells is independent of target cell density and averages 55 cells/mL. 10% purity can be achieved with as low as 6 CTCs/mL and better than 1% purity can be achieved with 1 CTC/mL.
Conclusions/significance: The LiquidBiopsy platform is an automated validated platform that provides high throughput molecular access to the CTC population. It can be validated and integrated into the lab flow enabling CTC enumeration as well as recovery of consistently high purity samples for molecular analysis such as quantitative PCR and Next Generation Sequencing. This tool opens the way for clinically relevant genetic profiling of CTCs.
Conflict of interest statement
Competing Interests: All authors are employees of Cynvenio Biosystems. This study was partly funded by Cynvenio Biosystems. PWD and PP are inventors of US patent 8,263,387, “Sheath flow devices and methods”, and BV and PD are inventors of US patent applications US2013/059251, “Spin Elute Tube”, both of which are related to the LiquidBiopsy® system. Cynvenio Biosystems has a commercial interest in the LiquidBiopsy® system. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Figures
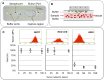
Figure 1. CTC Flow cell operation and performance.
(A) The LiquidBiopsy CTC flow cell. (B) A cartoon illustrating how target cells are pulled from the sheath flow while non targets move through the flow cell unhindered. (C) Efficiency of recovery of target cells using EpCAM based recovery: Incremental numbers of MCF7 (N = 34), HCC1419 (N = 27) or A549 (N = 65) cells were spiked into NHD blood and purified on the CTC flow cell. Recovered cells were enumerated in the flow cell. Graphs show number of targets spiked per mL of blood against the yield +/−1 SD from an average of between 7 and 27 experiments.
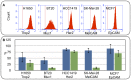
Figure 2. CTC Flow cell operation and performance.
(A) Flow cytometry histograms of H1650, BT20, H1419, SK-Mel-28 and MCF7 cells labeled with 1 ug/mL of anti Trop2, Muc1, Her2, MelCAM and EpCAM antibodies respectively. Control antibody threshold is indicated by the red line (B) Recovery and purity of target cells using magnetophoretic capture from NHD blood: 90 cells/mL were spiked into NHD blood and purified on the LiquidBiopsy platform. Recovered cells were enumerated in the flow cell. Graphs show average recovery and purity of target cells +/−1 SD from 4 experiments for H1650, BT20, H1419 and SK-Mel-28 capture and 100 experiments for MCF7 capture.
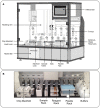
Figure 3. LiquidBiopsy automated platform.
(A) Diagram of the LiquidBiopsy platform. (B) Closeup of the platform worksurface. A X-Y pipetting arm with 4 pipettor heads transfers sample, buffers and antibody stains into 4 flow cells in the manifold. Sheath buffers are controlled by pumps on the rear wall of the platform.
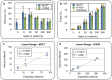
Figure 4. Validation of Automated CTC flow cell operation on LiquidBiopsy platform.
(A) Plot of CTC recovery as a function of spike-in density for MCF7 and H1650 cells. (B) Plot of sample purity as a function of spike in density for MCF7 and H1650 cells. The orange dots indicate the predicted purity of MCF7 cells if the non target recovery is held constant at 55 cells/mL. (C) Raw data and linear fit to MCF7 recovery curve from 9 to 90 cells/mL. (D) Raw data and linear fit to H1650 recovery curve from 9 to 300 cells/mL.
Figure 5. Direct molecular detection of tumor cells recovered from blood.
Results of a TaqMan PCR mutation assay for the EGFR Exon 19 deletion mutation in H1650 cells. DNA recovered from incremental numbers of H1650 tumor cells spiked into whole blood and processed on the LiquidBiopsy platform are indicated. The mutation is undetectable in whole blood even at the highest input.
Similar articles
- Nanoroughened adhesion-based capture of circulating tumor cells with heterogeneous expression and metastatic characteristics.
Chen W, Allen SG, Reka AK, Qian W, Han S, Zhao J, Bao L, Keshamouni VG, Merajver SD, Fu J. Chen W, et al. BMC Cancer. 2016 Aug 8;16:614. doi: 10.1186/s12885-016-2638-x. BMC Cancer. 2016. PMID: 27501846 Free PMC article. - EpCAM-independent capture of circulating tumor cells with a 'universal CTC-chip'.
Chikaishi Y, Yoneda K, Ohnaga T, Tanaka F. Chikaishi Y, et al. Oncol Rep. 2017 Jan;37(1):77-82. doi: 10.3892/or.2016.5235. Epub 2016 Nov 8. Oncol Rep. 2017. PMID: 27840987 - [Recent advances in isolation and detection of circulating tumor cells with a microfluidic system].
Cao R, Zhang M, Yu H, Qin J. Cao R, et al. Se Pu. 2022 Mar 8;40(3):213-223. doi: 10.3724/SP.J.1123.2021.07009. Se Pu. 2022. PMID: 35243831 Free PMC article. Review. Chinese. - A negative selection methodology using a microfluidic platform for the isolation and enumeration of circulating tumor cells.
Casavant BP, Mosher R, Warrick JW, Maccoux LJ, Berry SM, Becker JT, Chen V, Lang JM, McNeel DG, Beebe DJ. Casavant BP, et al. Methods. 2013 Dec 1;64(2):137-43. doi: 10.1016/j.ymeth.2013.05.027. Epub 2013 Jun 24. Methods. 2013. PMID: 23806645 Free PMC article. - Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells.
Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Lin M, et al. Acc Chem Res. 2014 Oct 21;47(10):2941-50. doi: 10.1021/ar5001617. Epub 2014 Aug 11. Acc Chem Res. 2014. PMID: 25111636 Free PMC article. Review.
Cited by
- Using circulating tumor cells to inform on prostate cancer biology and clinical utility.
Li J, Gregory SG, Garcia-Blanco MA, Armstrong AJ. Li J, et al. Crit Rev Clin Lab Sci. 2015;52(4):191-210. doi: 10.3109/10408363.2015.1023430. Epub 2015 Jun 16. Crit Rev Clin Lab Sci. 2015. PMID: 26079252 Free PMC article. Review. - Relevance of Circulating Tumor Cells as Predictive Markers for Cancer Incidence and Relapse.
Chelakkot C, Yang H, Shin YK. Chelakkot C, et al. Pharmaceuticals (Basel). 2022 Jan 6;15(1):75. doi: 10.3390/ph15010075. Pharmaceuticals (Basel). 2022. PMID: 35056131 Free PMC article. Review. - Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities.
Habli Z, AlChamaa W, Saab R, Kadara H, Khraiche ML. Habli Z, et al. Cancers (Basel). 2020 Jul 17;12(7):1930. doi: 10.3390/cancers12071930. Cancers (Basel). 2020. PMID: 32708837 Free PMC article. Review. - Analysis of Circulating Tumor Cells in Patients with Non-Metastatic High-Risk Prostate Cancer before and after Radiotherapy Using Three Different Enumeration Assays.
Budna-Tukan J, Świerczewska M, Mazel M, Cieślikowski WA, Ida A, Jankowiak A, Antczak A, Nowicki M, Pantel K, Azria D, Zabel M, Alix-Panabières C. Budna-Tukan J, et al. Cancers (Basel). 2019 Jun 10;11(6):802. doi: 10.3390/cancers11060802. Cancers (Basel). 2019. PMID: 31185699 Free PMC article. - Circulating Tumor Cells as a Marker of Disseminated Disease in Patients with Newly Diagnosed High-Risk Prostate Cancer.
Cieślikowski WA, Budna-Tukan J, Świerczewska M, Ida A, Hrab M, Jankowiak A, Mazel M, Nowicki M, Milecki P, Pantel K, Alix-Panabières C, Zabel M, Antczak A. Cieślikowski WA, et al. Cancers (Basel). 2020 Jan 9;12(1):160. doi: 10.3390/cancers12010160. Cancers (Basel). 2020. PMID: 31936460 Free PMC article.
References
- Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, et al. (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical Cancer Research 10: 6897–6904. - PubMed
- Fiegl M, Kircher B, Zojer N (2003) Correspondence re: T. Fehm et al., Cytogenetic Evidence that Circulating Epithelial Cells in Patients with Carcinoma Are Malignant. Clin. Cancer Res., 8: 2073–2084, 2002. Clinical Cancer Research 9: 1224–1225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous