PD-1 identifies the patient-specific CD8⁺ tumor-reactive repertoire infiltrating human tumors - PubMed (original) (raw)
Clinical Trial
. 2014 May;124(5):2246-59.
doi: 10.1172/JCI73639. Epub 2014 Mar 25.
Paul F Robbins, Xin Yao, Yong F Li, Simon Turcotte, Eric Tran, John R Wunderlich, Arnold Mixon, Shawn Farid, Mark E Dudley, Ken-Ichi Hanada, Jorge R Almeida, Sam Darko, Daniel C Douek, James C Yang, Steven A Rosenberg
- PMID: 24667641
- PMCID: PMC4001555
- DOI: 10.1172/JCI73639
Clinical Trial
PD-1 identifies the patient-specific CD8⁺ tumor-reactive repertoire infiltrating human tumors
Alena Gros et al. J Clin Invest. 2014 May.
Abstract
Adoptive transfer of tumor-infiltrating lymphocytes (TILs) can mediate regression of metastatic melanoma; however, TILs are a heterogeneous population, and there are no effective markers to specifically identify and select the repertoire of tumor-reactive and mutation-specific CD8⁺ lymphocytes. The lack of biomarkers limits the ability to study these cells and develop strategies to enhance clinical efficacy and extend this therapy to other malignancies. Here, we evaluated unique phenotypic traits of CD8⁺ TILs and TCR β chain (TCRβ) clonotypic frequency in melanoma tumors to identify patient-specific repertoires of tumor-reactive CD8⁺ lymphocytes. In all 6 tumors studied, expression of the inhibitory receptors programmed cell death 1 (PD-1; also known as CD279), lymphocyte-activation gene 3 (LAG-3; also known as CD223), and T cell immunoglobulin and mucin domain 3 (TIM-3) on CD8⁺ TILs identified the autologous tumor-reactive repertoire, including mutated neoantigen-specific CD8⁺ lymphocytes, whereas only a fraction of the tumor-reactive population expressed the costimulatory receptor 4-1BB (also known as CD137). TCRβ deep sequencing revealed oligoclonal expansion of specific TCRβ clonotypes in CD8⁺PD-1⁺ compared with CD8⁺PD-1- TIL populations. Furthermore, the most highly expanded TCRβ clonotypes in the CD8⁺ and the CD8⁺PD-1⁺ populations recognized the autologous tumor and included clonotypes targeting mutated antigens. Thus, in addition to the well-documented negative regulatory role of PD-1 in T cells, our findings demonstrate that PD-1 expression on CD8⁺ TILs also accurately identifies the repertoire of clonally expanded tumor-reactive cells and reveal a dual importance of PD-1 expression in the tumor microenvironment.
Figures
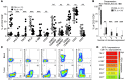
Figure 1. CD8+ TILs exhibit unique phenotypic traits compared with PBLs.
(A) Phenotypic characterization of CD8+ PBLs and TILs in melanoma patients. Percentages of PBLs and TILs expressing individual or combination of markers are shown (mean ± SEM). Each dot represents 1 sample analyzed. **P < 0.01, ***P < 0.001, Mann-Whitney test. (B) Coexpression of PD-1, LAG-3, TIM-3, and 4-1BB in CD8+ PBLs and TILs. The frequency of cells expressing 0, 1, 2, 3, or 4 markers is shown. Bars represent maximum, minimum, and mean values. **P < 0.01, ***P < 0.001, Mann-Whitney test. (C) Coexpression pattern of PD-1 and LAG-3, TIM-3, 4-1BB, CD27, and CD57 on CD8+ PBLs and TILs. Dot plots display the phenotype of CD8+ lymphocytes from matched samples from 1 representative patient. The percentage of cells expressing each combination of receptors is shown. (D) Frequency of PD-1 expression on CD8+ TIL subpopulations. TIM-3+, LAG-3+, 4-1BB+, CD25+, BTLA+, CD57+, and CD27+ CD8+ TILs were gated, and the frequency of PD-1 expression within these populations is summarized. Markers are displayed in order and color coded according to frequency of PD-1 expression. Mean ± SEM are shown. *P < 0.005 vs. BTLA+, CD57+, and CD27+, Dunn test for multiple comparisons.
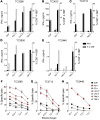
Figure 2. PD-1+, LAG-3+, and TIM-3+ derived CD8+ TILs, but not the negative counterparts, recognize and lyse their autologous tumor cell line.
Bulk CD3+CD8+ TILs were sorted to high purity from 6 tumors based on positive or negative expression of PD-1, LAG-3, and TIM-3 and expanded in vitro. (A) Response of FrTu1913-derived TILs to TC1913. TILs were cocultured with autologous TC1913, and tumor recognition was assessed by measuring IFN-γ release (duplicates, mean ± SD), and the frequency of CD8+4-1BB+ cells. (B) Reactivity of PD-1+ and PD-1– CD8+ TILs derived from FrTu1913 against a panel of targets: TC1913 with and without HLA-I blocking antibody (W6/32), TC2448 (matched HLA-A*0201), TC2301 (allogeneic), and plate-bound anti-CD3 (OKT3). Upregulation of 4-1BB (top) and IFN-γ release (bottom, duplicates, mean ± SD) are shown. (C) Lysis of TC1913 by FrTu1913-derived TILs. Cytotoxicity of TILs against TC1913 (mean ± SD). (D) Response of PD-1– and PD-1+ derived TILs to their autologous tumor targets from all the 6 fresh tumors studied. PD-1– and PD-1+ TILs were cocultured with their autologous tumor cell lines, and reactivity was assessed by measuring IFN-γ secretion (left) and 4-1BB upregulation (right). Each dot represents 1 patient’s sample. Mean ± SEM. *P ≤ 0.05, 2-tailed Wilcoxon signed-rank test.
Figure 3. Recognition and lysis of autologous tumor by CD8+ TILs sorted based on PD-1, LAG-3, and TIM-3 expression.
Bulk CD3+CD8+ TILs were sorted to high purity from FrTu3289, FrTu3612, FrTu3713, FrTu3550, and FrTu2448 based on positive or negative expression of PD-1, LAG-3 and/or TIM-3, and expanded in vitro for 15 days. (A–E) Response of fresh tumor–derived TILs to their respective autologous tumor cell lines, TC3289 (A), TC3612 (B), TC3713 (C), TC3550 (D) and TC2448 (E). Reactivity was assessed by measuring IFN-γ release (duplicates, mean ± SD) and frequency of 4-1BB upregulation. (F–H) Cytolytic activity of fresh tumor–derived TILs in response to their respective autologous tumor cell lines, TC3289 (F), TC3713 (G), and TC2448 (H). Percentage of specific lysis at different effector/target ratios is shown as mean ± SD.
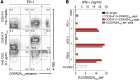
Figure 4. PD-1+, LAG-3+, and TIM-3+ derived CD8+ TILs prospectively identify tumor-reactive CD8+ TILs targeting a mutation in the CDKN2A gene locus.
CD3+CD8+ TILs were sorted to high purity from FrTu1913 based on positive or negative expression of PD-1, LAG-3, and TIM-3 and expanded in vitro. (A) Recognition of _CDKN2A_mut peptide pulsed COS-A11 cells by PD-1+ and PD-1– TILs. TILs were cocultured with COS-A11 cells pulsed with an irrelevant A*1101 restricted peptide, with _CDKN2A_mut peptide (1 μM), or with plate-bound anti-CD3. Dot plots display the frequency of expression of 4-1BB and _CDKN2A_mut peptide HLA-A*1101 tetramer complex binding of PD-1– (left) and PD-1+ (right) derived CD8+ TILs 24 hours after coculture. (B) IFN-γ secretion and frequency of _CDKN2A_mut tetramer+ cells in FrTu1913-derived TILs. TILs derived from FrTu1913 and control TIL3309, recognizing _CRKRS_mut HLA-A*1101 peptide, were cocultured with COS-A11 alone or pulsed with specific or irrelevant peptides; mean secretion of IFN-γ (duplicates) is represented. The frequency of _CDKN2A_mut-specific cells in the TIL populations was determined using a _CDKN2A_mut peptide HLA-A*1101 tetramer complex after gating on CD3+CD8+ cells (gray bars).
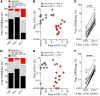
Figure 5. Expression of PD-1, rather than 4-1BB, more comprehensively identifies the repertoire of tumor-reactive cells in human tumors.
(A) Representative dot plots showing PD-1 by 4-1BB expression of CD8+ TILs infiltrating FrTu1913 and FrTu3713. (B) Expression of PD-1 by 4-1BB on CD8+ TILs infiltrating melanoma tumors. The frequency of each combination of markers is shown as mean ± SEM. ***P < 0.001, Wilcoxon signed-rank test (n = 24). (C) Response of FrTu1913-derived clones to TC1913. CD8+ TILs were sorted from FrTu1913 into PD-1–/4-1BB–, PD-1+/4-1BB–, and PD-1+/4-1BB+, and clones were established. Clones derived from PD-1–/4-1BB– (n = 96), PD-1+/4-1BB– (n = 57), and PD-1+/4-1BB+ (n = 65) were cocultured with TC1913; 4-1BB upregulation upon coculture is plotted for each clone. The line represents the median. *P ≤ 0.05, ****P ≤ 0.0001, Dunn test for multiple comparisons. Pie charts depict the percentage of tumor-reactive and non–tumor-reactive clones in each population. Greater than 1% CD8+4-1BB+ and greater than twice the background percentage 4-1BB compared with no stimulation control was considered positive. (D) Response of bulk-expanded FrTu3713-derived TILs to TC3713. CD8+PD-1–/4-1BB–, PD-1+/4-1BB–, and PD-1+/4-1BB+ TILs were sorted from FrTu3713 and expanded. Cells were either left unstimulated, cocultured with TC3713, or stimulated with plate-bound anti-CD3. 4-1BB upregulation was measured to assess tumor recognition. Representative dot plots of CD3+CD8+-gated cells are shown. (E) Lysis of TC3713 by TILs derived from FrTu3713.
Figure 6. Skewed repertoire and binary distribution of CD8+ TIL TCRβ clonotypes based on expression of PD-1 in the tumor.
CD8+ T cells as well as PD-1+ and PD-1– fractions were sorted to high purity from FrTu1913 (A–C) and FrTu3713 (D–F), mRNA was extracted, and TCRβ deep sequencing was performed. (A and D) Diversity of the TCRβ repertoire within the bulk CD8+, CD8+PD-1–, and CD8+PD-1+ TIL populations. The relative frequencies of the most frequent TCRβ clonotype (unique CDR3β amino acid sequences), the second most frequent, the 3rd to 30th most frequent, and the rest of the clonotypes are shown. (B and E) The 30 most frequent CD8+ TIL clonotypes in the fresh tumor are plotted based on their frequency in PD-1+ and PD-1– populations. (C and F) Frequency of the 30 most frequent TCRβ clonotypes in the CD8+PD-1+ TIL population. Their frequency in the PD-1+ and PD-1– subsets is represented. Each dot represents 1 unique TCRβ clonotype. The cumulative frequency (∑ freq.) of the clonotypes in each of the populations is shown below. ****P ≤ 0.0001, Wilcoxon signed-rank test.
Figure 7. Tumor-reactive and mutation-specific clonotypes are highly expanded in the CD8+ and PD-1+ populations.
(A) Response of CD8+PD-1+ (n = 44) and CD8+PD-1– (n = 109) FrTu3713-derived T cell clones to TC3713. Each dot represents 1 clone; line represents median IFN-γ release for all clones tested. The percentage of tumor-reactive clones (>50 pg/ml IFN-γ) is shown above. ****P ≤ 0.0001, Mann-Whitney test. (B) Reactivity of FrTu1913-derived clones against autologous tumor TC1913 or allogeneic TC624 (matched HLA-A*0201). Clone 41 expressed the CDR3β amino acid sequence corresponding to the most frequent CD8+ clonotype in FrTu1913; clone 208 recognized _CDKN2A_mut HLA-A*1101 restricted epitope; clone 199 was a tumor-reactive clone of yet-unknown specificity; clone 88 did not recognize TC1913. Recognition of targets was measured by assessing 4-1BB upregulation. (C) Recognition of _CDKN2A_mut and HLA-A11mut by FrTu1913-derived T cell clones. COS-A2 cells were transiently transfected with plasmids encoding HLA-A11WT or HLA-A11mut, and recognition of _CDKN2A_mut peptide was assessed by measuring 4-1BB upregulation. (D) Frequency of the 30 most frequent TCRβ clonotypes in the CD8+PD-1+ population in FrTu1913. The cumulative frequency (Σ freq.) of the HLA-A11mut–specific clonotypes in each population is specified below.
Similar articles
- Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression.
Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, Rosenberg SA. Parkhurst M, et al. Clin Cancer Res. 2017 May 15;23(10):2491-2505. doi: 10.1158/1078-0432.CCR-16-2680. Epub 2016 Nov 8. Clin Cancer Res. 2017. PMID: 27827318 Free PMC article. - Co-stimulation through 4-1BB/CD137 improves the expansion and function of CD8(+) melanoma tumor-infiltrating lymphocytes for adoptive T-cell therapy.
Chacon JA, Wu RC, Sukhumalchandra P, Molldrem JJ, Sarnaik A, Pilon-Thomas S, Weber J, Hwu P, Radvanyi L. Chacon JA, et al. PLoS One. 2013;8(4):e60031. doi: 10.1371/journal.pone.0060031. Epub 2013 Apr 1. PLoS One. 2013. PMID: 23560068 Free PMC article. - PD-1+ Polyfunctional T Cells Dominate the Periphery after Tumor-Infiltrating Lymphocyte Therapy for Cancer.
Donia M, Kjeldsen JW, Andersen R, Westergaard MCW, Bianchi V, Legut M, Attaf M, Szomolay B, Ott S, Dolton G, Lyngaa R, Hadrup SR, Sewell AK, Svane IM. Donia M, et al. Clin Cancer Res. 2017 Oct 1;23(19):5779-5788. doi: 10.1158/1078-0432.CCR-16-1692. Epub 2017 Jul 5. Clin Cancer Res. 2017. PMID: 28679768 Free PMC article. Clinical Trial. - Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy.
Durgeau A, Virk Y, Corgnac S, Mami-Chouaib F. Durgeau A, et al. Front Immunol. 2018 Jan 22;9:14. doi: 10.3389/fimmu.2018.00014. eCollection 2018. Front Immunol. 2018. PMID: 29403496 Free PMC article. Review. - Adoptive T-cell therapy using autologous tumor-infiltrating lymphocytes for metastatic melanoma: current status and future outlook.
Wu R, Forget MA, Chacon J, Bernatchez C, Haymaker C, Chen JQ, Hwu P, Radvanyi LG. Wu R, et al. Cancer J. 2012 Mar-Apr;18(2):160-75. doi: 10.1097/PPO.0b013e31824d4465. Cancer J. 2012. PMID: 22453018 Free PMC article. Review.
Cited by
- Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells.
Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH, Mardis ER, Linette GP. Carreno BM, et al. Science. 2015 May 15;348(6236):803-8. doi: 10.1126/science.aaa3828. Epub 2015 Apr 2. Science. 2015. PMID: 25837513 Free PMC article. Clinical Trial. - Tissue-resident memory-like T cells in tumor immunity: Clinical implications.
Dhodapkar MV, Dhodapkar KM. Dhodapkar MV, et al. Semin Immunol. 2020 Jun;49:101415. doi: 10.1016/j.smim.2020.101415. Epub 2020 Sep 30. Semin Immunol. 2020. PMID: 33011063 Free PMC article. Review. - Expression of Galectin-9-related immune checkpoint receptors in B-cell acute lymphoblastic leukemia.
Akbar A, Asgarian-Omran H, Valadan R, Dindarloo MM, Najafi A, Kahrizi A, Poursheikhani A, Karami H, Naderi M, Sabeti S, Tehrani M. Akbar A, et al. Iran J Basic Med Sci. 2023;26(12):1468-1474. doi: 10.22038/IJBMS.2023.73159.15901. Iran J Basic Med Sci. 2023. PMID: 37970435 Free PMC article. - Synthetic TILs: Engineered Tumor-Infiltrating Lymphocytes With Improved Therapeutic Potential.
Jiménez-Reinoso A, Nehme-Álvarez D, Domínguez-Alonso C, Álvarez-Vallina L. Jiménez-Reinoso A, et al. Front Oncol. 2021 Feb 16;10:593848. doi: 10.3389/fonc.2020.593848. eCollection 2020. Front Oncol. 2021. PMID: 33680923 Free PMC article. Review. - Reversing T-cell Dysfunction and Exhaustion in Cancer.
Zarour HM. Zarour HM. Clin Cancer Res. 2016 Apr 15;22(8):1856-64. doi: 10.1158/1078-0432.CCR-15-1849. Clin Cancer Res. 2016. PMID: 27084739 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials