Degenerate target sites mediate rapid primed CRISPR adaptation - PubMed (original) (raw)
Degenerate target sites mediate rapid primed CRISPR adaptation
Peter C Fineran et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Prokaryotes encode adaptive immune systems, called CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated), to provide resistance against mobile invaders, such as viruses and plasmids. Host immunity is based on incorporation of invader DNA sequences in a memory locus (CRISPR), the formation of guide RNAs from this locus, and the degradation of cognate invader DNA (protospacer). Invaders can escape type I-E CRISPR-Cas immunity in Escherichia coli K12 by making point mutations in the seed region of the protospacer or its adjacent motif (PAM), but hosts quickly restore immunity by integrating new spacers in a positive-feedback process termed "priming." Here, by using a randomized protospacer and PAM library and high-throughput plasmid loss assays, we provide a systematic analysis of the constraints of both direct interference and subsequent priming in E. coli. We have defined a high-resolution genetic map of direct interference by Cascade and Cas3, which includes five positions of the protospacer at 6-nt intervals that readily tolerate mutations. Importantly, we show that priming is an extremely robust process capable of using degenerate target regions, with up to 13 mutations throughout the PAM and protospacer region. Priming is influenced by the number of mismatches, their position, and is nucleotide dependent. Our findings imply that even outdated spacers containing many mismatches can induce a rapid primed CRISPR response against diversified or related invaders, giving microbes an advantage in the coevolutionary arms race with their invaders.
Keywords: adaptive immunity; crRNA; horizontal gene transfer; next-generation sequencing; phage resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
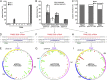
Fig. 1.
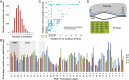
Preexisting spacers with up to seven mismatches promote priming. (A) Loss of plasmid pGFPuv from Δ_hns_ and various pRSF-1b PIM derivatives (20). (B) Plasmid loss in Δ_hns_, PIM25, and PIM2 backgrounds for plasmids pGFPuv and pACYC184. The percentage of plasmid-free clones containing no spacers (white) or at least one new spacer (gray) is shown. (C) Percentage of spacers derived from forward (priming) or reverse strands of the plasmids from B. (D) Match of PIM25 S26 crRNA to the protospacer in pGFPuv. (E) Mapping of new spacers acquired by PIM25 following loss of pGFPuv. (F) Match of PIM25 S26 crRNA to the protospacer in pACYC184. (G) Mapping of new spacers acquired by PIM25 following loss of pACYC184. (H) Match of PIM2 S22 crRNA to the protospacer in pGFPuv. (I) Mapping of new spacers acquired by PIM2 following loss of pGFPuv. In D, F, and H the spacer (red), protospacer (blue), PAM (green), and mismatches (bold) are indicated. In E, G, and I the protospacer (PS) region is indicated in purple, new forward (primed) spacers in pale green, new reverse spacers in red, and all of the respective consensus PAMs are shown in green (forward) and orange (reverse).
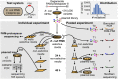
Fig. 2.
Experimental design for high-throughput individual and pooled plasmid-loss experiments. A test system of E. coli PIM5 containing PS8 was selected that would target a pGFPuv-Km plasmid containing a consensus PAM and PS8. A degenerate PAM-PS8 library of variants was generated in pGFPuv-Km, with an average distribution of five mutations per insert (histogram). For the individual experiment, the plasmid library was transformed into PIM5, individual colonies were sequenced, plasmid-loss experiments without selection performed, and a subset of variants checked for spacer acquisition by PCR and sequencing. In the pooled experiment, plasmid DNA was prepared for the original library (T0), which was then transformed into PIM5 (T1) and then passaged for plasmid loss without selection for 24 h (T2) and 48 h (T3). For T0–T3, samples were amplified with barcoded primers, pooled, and sequenced.
Fig. 3.
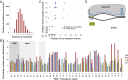
Up to 11 mutations within the protospacer and PAM region promote priming. (A) Plasmid loss at 48 h for 366 individual loss experiments relative to the number of mutations in each protospacer and PAM (lines represent average loss for variants with that number of mutations). (B) Twenty variants with between 1 and 11 mutations that displayed plasmid loss via priming. The spacers acquired by these variants are shown in C and details are in
Table S4
and
Datasets S1
and
S2
.
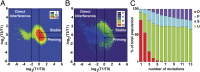
Fig. 4.
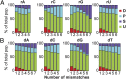
Classification of functional behavior of individual PAM-protospacer variants in the pooled loss experiment. (A) Contour map of the number of variants. The depletion ratio of the number of reads at T1 and T0 (revealing direct interference) is plotted on a double log2 scale vs. the ratio of reads at T3 and T1 (revealing priming). Variants were binned in 0.1 by 0.1 bins and the number of sequences per bin is shown in a color range of blue to red (1–100 variants). Black boxes indicate the boundaries of the three different functional categories [direct interference (x < −2, _n_ = 8,792), priming (_x_ > 0, y < −0.5, _n_ = 26,842), and stable (_x_ > 0, y > 0.5, n = 12,066)] and a group of unclassified (n = 86,395) variants. (B) Contour map as in A, showing the average number of mutations of each bin in a color range of purple to red (0–12 mutations). (C) Percentage distribution of the functional categories at increasing numbers of mutations (D, direction interference; P, priming; S, stable; U, unclassified).
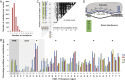
Fig. 5.
Analysis of variants displaying direct interference. (A) Distribution of the number of mutations per variant. (B) Percentage mutation per substitution type per position. For example, ∼10% of all G-to-A mutations at position 6 end up in the direct interference class. Asterisks highlight the position 6, 12, 18, 24, and 30 pinch points of Cascade. (C) Analysis of the mutation position of all double mutants (n = 2,791 sequences) found in the direct interference class. Protospacer mutations were scored regardless of the identity of the nucleotide, whereas PAM nucleotides were scored taking the nucleotide identity into account. Bubble size is indicative of the fraction of variants with mutations at a certain combination of positions displaying direct interference behavior. The absence of a bubble indicates that a certain combination of mutations leads to escape from direct interference. (D) Schematic representation of five PAM sequences on the target strand from 5′ to 3′ supporting direct interference (see also
Table S7
). The five pinch point positions of Cas7 where mutations are readily tolerated for direct interference (red base pairs numbered 6, 12, 18, 24, and 30), as well as the five helical segments within the Cascade R-loop (gray lines, numbered 1–5) are indicated.
Fig. 6.
Analysis of variants displaying priming. (A) Distribution of the number of mutations per variant. (B) Percentage mutation per substitution type per position. For example, ∼40% of all C-to-A mutations at position 28 end up in the priming class. (C) Analysis of pairs of mutations in triple mutants that significantly (P < 0.05) contribute to priming behavior (n = 17,109 triple-mutant sequences). Protospacer mutations were scored regardless of the identity of the nucleotide, whereas PAM nucleotides were scored taking the nucleotide identity into account. The absence of a point indicates that a certain combination of mutations does not significantly lead to priming. (D) Schematic representation of the Cascade R-loop indicating PAM sequences on the target strand from 5′ to 3′ supporting priming (see also
Table S7
). PAM sequences indicated with an asterisk (*) were computationally inferred from the analysis of the behavior of the sequences containing the given PAM and either one or two additional mutations (see
Table S7
). Protospacer position 28, which is highly associated with priming, is shown in red.
Fig. 7.
Analysis of stable variants. (A) Distribution of the number of mutations per variant. (B) Percentage mutation per substitution type per position. For example, ∼20% of all T-to-C mutations at positions 29, 30, and 31 end up in the stable class. (C) Analysis of pairs of mutations in triple mutants that significantly (P < 0.05) contribute to stable behavior (n = 17,109 triple-mutant sequences). Protospacer mutations were scored regardless of the identity of the nucleotide, whereas PAM nucleotides were scored taking the nucleotide identity into account. The absence of a point indicates that a certain combination of mutations does not significantly lead to priming. (D) Schematic representation of the Cascade R-loop indicating that no PAM sequences are sufficient to cause stable behavior. Positions of variants overrepresented in the stable class are shown in red (positions 10–12, 18, 22, 24, 25) and orange (positions 29–31).
Fig. 8.
Effect of mismatches between the crRNA spacer sequence and the targeted strand of the protospacer. Variants with mutations in the PAM were excluded in this analysis (remaining group n = 83,655). (A) Mismatched ribonucleotides in the crRNA spacer. (B) Mismatched deoxyribonucleotides in the targeted strand of the protospacer DNA.
Similar articles
- CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli.
Díez-Villaseñor C, Guzmán NM, Almendros C, García-Martínez J, Mojica FJ. Díez-Villaseñor C, et al. RNA Biol. 2013 May;10(5):792-802. doi: 10.4161/rna.24023. Epub 2013 Feb 27. RNA Biol. 2013. PMID: 23445770 Free PMC article. - Cas3 is a limiting factor for CRISPR-Cas immunity in Escherichia coli cells lacking H-NS.
Majsec K, Bolt EL, Ivančić-Baće I. Majsec K, et al. BMC Microbiol. 2016 Mar 8;16:28. doi: 10.1186/s12866-016-0643-5. BMC Microbiol. 2016. PMID: 26956996 Free PMC article. - Avoidance of Trinucleotide Corresponding to Consensus Protospacer Adjacent Motif Controls the Efficiency of Prespacer Selection during Primed Adaptation.
Musharova O, Vyhovskyi D, Medvedeva S, Guzina J, Zhitnyuk Y, Djordjevic M, Severinov K, Savitskaya E. Musharova O, et al. mBio. 2018 Dec 4;9(6):e02169-18. doi: 10.1128/mBio.02169-18. mBio. 2018. PMID: 30514784 Free PMC article. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - Methods for decoding Cas9 protospacer adjacent motif (PAM) sequences: A brief overview.
Karvelis T, Gasiunas G, Siksnys V. Karvelis T, et al. Methods. 2017 May 15;121-122:3-8. doi: 10.1016/j.ymeth.2017.03.006. Epub 2017 Mar 24. Methods. 2017. PMID: 28344037 Review.
Cited by
- Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa.
Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B. Rollins MF, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2216-22. doi: 10.1093/nar/gkv094. Nucleic Acids Res. 2015. PMID: 25662606 Free PMC article. - The diversity-generating benefits of a prokaryotic adaptive immune system.
van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, Westra ER. van Houte S, et al. Nature. 2016 Apr 21;532(7599):385-8. doi: 10.1038/nature17436. Epub 2016 Apr 13. Nature. 2016. PMID: 27074511 Free PMC article. - CRISPR-Cas adaptation in Escherichia coli.
Mitić D, Bolt EL, Ivančić-Baće I. Mitić D, et al. Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review. - CRISPR analysis suggests that small circular single-stranded DNA smacoviruses infect Archaea instead of humans.
Díez-Villaseñor C, Rodriguez-Valera F. Díez-Villaseñor C, et al. Nat Commun. 2019 Jan 17;10(1):294. doi: 10.1038/s41467-018-08167-w. Nat Commun. 2019. PMID: 30655519 Free PMC article. - Adaptation by Type III CRISPR-Cas Systems: Breakthrough Findings and Open Questions.
Zhang X, An X. Zhang X, et al. Front Microbiol. 2022 Apr 14;13:876174. doi: 10.3389/fmicb.2022.876174. eCollection 2022. Front Microbiol. 2022. PMID: 35495695 Free PMC article. Review.
References
- Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: Defeating bacterial defences. Nat Rev Microbiol. 2013;11(10):675–687. - PubMed
- Westra ER, et al. The CRISPRs, they are a-changin’: How prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. - PubMed
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4(3):267–278. - PubMed
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous