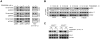Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors - PubMed (original) (raw)
. 2014 May 12;25(5):697-710.
doi: 10.1016/j.ccr.2014.03.011. Epub 2014 Apr 17.
Anna Saborowski 2, Jingyin Yue 3, Martha Solomon 3, Eric Joseph 3, Sunyana Gadal 3, Michael Saborowski 2, Edward Kastenhuber 2, Christof Fellmann 4, Kazuhiro Ohara 5, Kenji Morikami 5, Takaaki Miura 5, Christine Lukacs 6, Nobuya Ishii 5, Scott Lowe 7, Neal Rosen 8
Affiliations
- PMID: 24746704
- PMCID: PMC4049532
- DOI: 10.1016/j.ccr.2014.03.011
Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors
Piro Lito et al. Cancer Cell. 2014.
Abstract
MEK inhibitors are clinically active in BRAF(V600E) melanomas but only marginally so in KRAS mutant tumors. Here, we found that MEK inhibitors suppress ERK signaling more potently in BRAF(V600E), than in KRAS mutant tumors. To understand this, we performed an RNAi screen in a KRAS mutant model and found that CRAF knockdown enhanced MEK inhibition. MEK activated by CRAF was less susceptible to MEK inhibitors than when activated by BRAF(V600E). MEK inhibitors induced RAF-MEK complexes in KRAS mutant models, and disrupting such complexes enhanced inhibition of CRAF-dependent ERK signaling. Newer MEK inhibitors target MEK catalytic activity and also impair its reactivation by CRAF, either by disrupting RAF-MEK complexes or by interacting with Ser 222 to prevent MEK phosphorylation by RAF.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Figure 1. KRAS mutant tumors are less sensitive to allosteric MEK inhibitors than BRAFV600E tumors
(A) BRAFV600E and KRAS-mutant tumor cell lines were treated with increasing doses of PD0325901 for three days to determine the effect on proliferation. A representative example of three independent experiments (each performed in triplicate) is shown as means +/- SEM. (B-D) The indicated tumor cell lines were treated with increasing amounts of PD0325901 for three hours (B) or with 50 nM of PD0325901 for the indicated times (C). Lysates were assayed by immunoblotting to determine the level of MEK and ERK phosphorylation. The bands were quantified by densitometry and the pERK level after treatment was normalized to the pretreatment pERK level (D). A representative example of three independent experiments for each cell line is shown. (E, F) Fold change in shRNA abundance in DMSO-treated (E) or PD0325901 (F)-treated cells. Note the selective depletion in shRNAs targeting CRAF with PD0325901 treatment. shRNAs targeting Rpa3 or KRAS were used as positive controls, whereas those targeting luciferase or renilla were used as negative controls. Bars represent the mean fold change from an experiment performed in triplicate. (G) KRPC cells were transduced with the indicated CRAF shRNAs and lysates were subjected to immunoblotting to determine the knockdown in CRAF expression. (H) KRPC cells infected with four different CRAF shRNAs were treated with dox to induce CRAF knockdown and PD0325901 (+MEK inhibitor) or DMSO (-MEK inhibitor) for the indicated times. The data were normalized to the shRNA abundance in cells after 48 hr of treatment with dox. (I) KRPC cells were stably infected with the indicated dox-inducible CRAF shRNA followed by implantation in athymic mice. The mice were treated with MEK inhibitor PD0325901 in the presence or absence of dox. The effect of CRAF knockdown (+dox) in the ability of the MEK inhibitor to inhibit KRAS tumor growth is represented as mean +/-SEM (n=6) See also Figure S1.
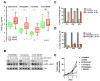
Figure 2. CRAF expression attenuates the effects of MEK inhibition in KRAS-mutant tumors
(A) KRAS-mutant lung cancer cells (A549) were transfected with siRNA pools targeting each of the RAF isoforms and then treated with 50 nM of PD0325901 for 48 hr. Whole cell lysates were evaluated by immunoblotting to determine the effect on ERK signaling. A representative example of three independent experiments for each siRNA is shown. (B) The indicated cell lines were treated with 50 nM of PD0325901 as shown. CRAF was immunoprecipitated from whole cell lysates and subjected to a kinase assay using an inactive MEK1 (K97R, KD) as the substrate. CRAF activity was determined by immunubloting with a phospho-MEK antibody. A representative example of two or more independent experiments for each cell line is shown. (C) Craf-/- MEFs were co-transfected with the indicated constructs and then treated with PD0325901 for 1 hr. Lysates were analyzed by immunobloting to determine ERK phosphorylation. A representative example of two independent experiments for each RAS isoform is shown. WT: wild type; KD: kinase dead See also Figure S2
Figure 3. MEK is less susceptible to inhibition when activated by CRAF than BRAF V600E
(A) A schematic representation of the experimental system used. BRAFV600E mutant A375 cells have low levels of CRAF activity at baseline and MEK activation is dependent on BRAF kinase. Treatment with RAF or MEK inhibitors inhibits ERK signaling in these cells. We relied on the ability of RAF inhibitors to transactivate RAF dimers in order to switch from BRAFV600E-dependent to catC-dependent signaling. To this end, catC was expressed to form BRAFV600E-catC heterodimers in A375 cells. A gatekeeper mutation in catC (T421M) prevents RAF inhibitor binding. Thus, the RAF inhibitor vemurafenib binds to and inhibits BRAFV600E while transactivating catC in heterodimers. A kinase-dead catC (T421M/K375M) was used as a control. (B) A375 cells expressing the indicated constructs were treated with increasing doses of vemurafenib for 1 hr and whole cell lysates were subjected to immunoblotting with the indicated antibodies. (C) A375 cells were transfected with increasing amounts of plasmid encoding catCT421M. In the indicated rows RAF inhibitor pre-treatment (vemurafenib, 1μM, 1 hr) was used to transactivate catC in heterodimers, and switch from BRAFV600E- to catC-driven signaling. Following transfection and pretreatment as shown, the cells were treated with increasing doses of PD0325901 for 1 hr to determine its ability to inhibit ERK phosphorylation. A representative example of at least two independent experiments for each condition is shown. (D, E) The bands in C were quantified by densitometry and normalized to the level of pERK prior to PD0325901 treatment. See also Figure S3
Figure 4. Allosteric MEK inhibitors induce RAF-MEK complex formation in KRAS mutant tumors
(A) HEK293 cells were transfected with FLAG-MEK1, followed by treatment with PD0325901 for 3 hrs. MEK1 was immunoprecipitated using an anti-FLAG antibody followed by immunoblotting with the indicated antibodies to determine RAF-MEK interactions. A representative example of at least two independent experiments with each drug is shown. (B) Whole cell lysates from A375 cells were subjected to immunoprecipitation with IgG or a MEK1 antibody and then immunoblotted for the indicated proteins. Two MEK specific IP replicates are shown. (C) Cells were treated with PD0325901 for 3 hrs and endogenous MEK1 was immunoprecipitated to determine its interaction with RAF kinases. A representative example of three independent experiments is shown. (D) HEK293 cells were transfected with WT MEK1 or a MEK1 mutant with impaired RAF interaction (IRI, M308A/I310A). RAF-MEK1 complexes were determined as in C. (E) A375 cells were transfected with the indicated constructs and then treated with PD0325901 (50 nM), selumetinib (500 nM) or RDEA119 (100 nM) for 1 hr to determine the effect on ERK phosphorylation. A representative example of at least two independent experiments for PD0325901 and selumetinib are shown. (F) A schematic diagram modeling the role of CRAF in the differential adaptation of KRAS and BRAF mutant tumors to MEK inhibitor treatment. See also Figure S4
Figure 5. Newer allosteric MEK inhibitors inhibit ERK phosphorylation better than PD0325901
(A) A panel of BRAFV600E (n, 15) and KRAS mutant (n, 50) tumor cell lines were subjected to proliferation assays with the indicated drugs. Cells were treated for three days with increasing amounts of the indicated compounds, in order to determine their respective IC50s. The mean (line) and range of IC50s across cell lines (n of 3 for each cell line) is shown. (B) KRAS mutant A549 cells were treated with increasing concentrations of PD0325901, trametinib or CH5126766 for 48 hr. Lysates were evaluated by immunoblotting to determine the level of ERK phosphorylation. A representative of two independent experiments is shown. (C, D) KRPC cells expressing dox-inducible CRAF shRNAs were treated with dox alone (no drug) or in combination with trametinib (C) or PD0325901 (D) at the indicated concentrations. The data were normalized to the number of shRNA expressing cells in the dox only controls. A representative of two independent experiments with at least two different shRNAs targeting CRAF is shown. (E) KRAS mutant A549 cells were treated with 100 nM PD0325901, 2 μM AZ628, or 100 nM trametinib as indicated. The fold increase in viable cell number over time is shown. Means +/- SEM from a representative of two independent experiments, each performed in triplicate, are shown. See also Figure S5
Figure 6. Trametinib binding weakens the interaction of MEK1 with RAF
(A) HEK293 cells were transfected with FLAG-MEK1 and then treated with PD0325901 (50 nM), trametinib (10 nM) or CH5126766 (250 nM) for 3 hr. MEK1 was immunoprecipitated with a FLAG antibody and RAF-MEK complexes were determined by immunoblotting. (B, C) BRAFV600E (A375, B) and KRASG12S (A549, C) cells were treated with the indicated MEK inhibitors as in (A). Lysates were subjected to immunoprecipitations and immunoblotting to determine the interaction between endogenous MEK1 and RAF kinases. (D) A549 cells were treated with PD0325901 (50 nM) or trametinib (10 nM) for the indicated times. RAF-MEK complexes were determined as in (C). (E) Recombinant GST-BRAF or GST-CRAF was immobilized on a sensor chip followed by sequential injections of increasing amounts of His-MEK1 in the presence or absence of trametinib at a saturating concentration (3 μM) to determine the association and dissociation rates of MEK-RAF complexes. A representative of at least two independent experiments is shown in each panel of this figure. (F) Effect of MEK inhibitors on the binding constants for the interaction between MEK1 and RAF. See also Figure S6
Figure 7. Interaction with Ser 222 and allosteric displacement of the MEK activation segment by CH5126766 prevents RAF-mediated phosphorylation
(A) The ternary structure of MEK1-bound to CH5126766 and an ATP analog at a resolution of 2.7 Å. Its interaction with the MEK activation segment residues Ser 212, Asn 221 and Ser 222 are shown. (B) Superpositioning of CH5126766 (green), CH4987655 (yellow) and PD0325089 (an enantiomer of PD0325901, red) and their effect on the orientation of the MEK1 activation segment. (C-E) Interactions of the indicated compounds with key residues in the MEK1 acitvation segment. (F) KRAS mutant A549 cells were treated with the indicated inhibitors for 3 hrs and whole cell lysates were subjected to immunoblotting with the indicated antibodies. A representative example of three independent experiments is shown. (G) HEK293 cells transfected with wild type MEK1 or a MEK1 mutant harboring phosphomimetic substitutions with glutamic acid at Ser 218 and Ser 222 (i.e. EE) were treated with CH5126766 for 3 hrs. Co-immunoprecipitations were used to determine the interaction between FLAG-tagged MEK1 and endogenous BRAF or CRAF. A representative example of two independent experiments is shown. (H) Schematic diagram representing the allosteric effects induced by each MEK inhibitor. See also Figure S7 and Table S1.
Similar articles
- RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth.
Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. Hatzivassiliou G, et al. Nature. 2010 Mar 18;464(7287):431-5. doi: 10.1038/nature08833. Epub 2010 Feb 3. Nature. 2010. PMID: 20130576 - RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF.
Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. Poulikakos PI, et al. Nature. 2010 Mar 18;464(7287):427-30. doi: 10.1038/nature08902. Nature. 2010. PMID: 20179705 Free PMC article. - Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib.
Acquaviva J, Smith DL, Jimenez JP, Zhang C, Sequeira M, He S, Sang J, Bates RC, Proia DA. Acquaviva J, et al. Mol Cancer Ther. 2014 Feb;13(2):353-63. doi: 10.1158/1535-7163.MCT-13-0481. Epub 2014 Jan 7. Mol Cancer Ther. 2014. PMID: 24398428 - Resistance to MEK inhibitors: should we co-target upstream?
Poulikakos PI, Solit DB. Poulikakos PI, et al. Sci Signal. 2011 Mar 29;4(166):pe16. doi: 10.1126/scisignal.2001948. Sci Signal. 2011. PMID: 21447797 Review. - MEK and RAF inhibitors for BRAF-mutated cancers.
Belden S, Flaherty KT. Belden S, et al. Expert Rev Mol Med. 2012 Oct 12;14:e17. doi: 10.1017/erm.2012.11. Expert Rev Mol Med. 2012. PMID: 23058743 Review.
Cited by
- Targeting Non-V600 Mutations in BRAF: A Single Institution Retrospective Analysis and Review of the Literature.
Chaudhary HA, Cannon TL, Winer A. Chaudhary HA, et al. Drugs R D. 2024 Sep;24(3):395-403. doi: 10.1007/s40268-024-00475-5. Epub 2024 Aug 23. Drugs R D. 2024. PMID: 39177935 Free PMC article. Review. - Drugging KRAS: current perspectives and state-of-art review.
Parikh K, Banna G, Liu SV, Friedlaender A, Desai A, Subbiah V, Addeo A. Parikh K, et al. J Hematol Oncol. 2022 Oct 25;15(1):152. doi: 10.1186/s13045-022-01375-4. J Hematol Oncol. 2022. PMID: 36284306 Free PMC article. Review. - Molecular inhibition of RAS signalling to target ageing and age-related health.
Laskovs M, Partridge L, Slack C. Laskovs M, et al. Dis Model Mech. 2022 Oct 1;15(10):dmm049627. doi: 10.1242/dmm.049627. Epub 2022 Sep 16. Dis Model Mech. 2022. PMID: 36111627 Free PMC article. Review. - The Predictive and Prognostic Role of RAS-RAF-MEK-ERK Pathway Alterations in Breast Cancer: Revision of the Literature and Comparison with the Analysis of Cancer Genomic Datasets.
Rocca A, Braga L, Volpe MC, Maiocchi S, Generali D. Rocca A, et al. Cancers (Basel). 2022 Oct 28;14(21):5306. doi: 10.3390/cancers14215306. Cancers (Basel). 2022. PMID: 36358725 Free PMC article. Review. - Drug resistance in targeted cancer therapies with RAF inhibitors.
Degirmenci U, Yap J, Sim YRM, Qin S, Hu J. Degirmenci U, et al. Cancer Drug Resist. 2021 Jun 17;4(3):665-683. doi: 10.20517/cdr.2021.36. eCollection 2021. Cancer Drug Resist. 2021. PMID: 35582307 Free PMC article. Review.
References
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews Drug discovery. 2012;11:873–886. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous