Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development - PubMed (original) (raw)
Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development
Sean C Bendall et al. Cell. 2014.
Abstract
Tissue regeneration is an orchestrated progression of cells from an immature state to a mature one, conventionally represented as distinctive cell subsets. A continuum of transitional cell states exists between these discrete stages. We combine the depth of single-cell mass cytometry and an algorithm developed to leverage this continuum by aligning single cells of a given lineage onto a unified trajectory that accurately predicts the developmental path de novo. Applied to human B cell lymphopoiesis, the algorithm (termed Wanderlust) constructed trajectories spanning from hematopoietic stem cells through to naive B cells. This trajectory revealed nascent fractions of B cell progenitors and aligned them with developmentally cued regulatory signaling including IL-7/STAT5 and cellular events such as immunoglobulin rearrangement, highlighting checkpoints across which regulatory signals are rewired paralleling changes in cellular state. This study provides a comprehensive analysis of human B lymphopoiesis, laying a foundation to apply this approach to other tissues and "corrupted" developmental processes including cancer.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
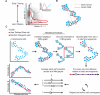
Figure 1. Developmental trajectory detection
A) Non-linear relationships between developmentally related cells. Markers ‘A’ and ‘B’ represent sequentially expressed phenotypic epitopes on cells in a developing system (inset). The red line shows the expected developmental trajectory from the earliest (cell ‘X’) to the most mature cell type (cell ‘Y’). Developmentally, the distant cell types can be close in Euclidean space. B) Determining the shortest path through a graph of the data reflects temporal distance between cells (solid red line between early (cell ‘X’) and target (cell ‘Y’)) better than standard metrics (e.g. Euclidian norm or correlation). Short circuits (dashed red line) impede a naïve shortest path-based algorithm. C) Description of the Wanderlust algorithm. The input data is single cells in N-dimensional space (top left). Wanderlust transforms the data into an ensemble of graphs and selects random waypoints (purple). Each graph is independently analyzed (single graph, red box) where a user-defined starting cell (red) is used to calculate an orientation trajectory. The orientation trajectory is iteratively refined using the waypoint cells. The final trajectory is an average over all graphs. To examine trends, the trace of each marker can be plotted according to trajectory position. See also Figure S1 for evaluation of Wanderlust on simulated data.
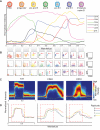
Figure 2. Wanderlust confirms known hallmarks of human B cell development and is consistent across healthy individuals
A) The Wanderlust trajectory is fixed to an arbitrary scale where the most immature cells are at 0 and the most mature cells at 1. The traces (based on median marker levels within a sliding window) demonstrate the relative expression patterns of CD34, CD38, CD10, CD19, IgH (s)urface, and CD20 across development. The approximate position of progenitors and B cell fractions is indicated. B) Biaxial plots demonstrate the two-dimensional progression of cellular marker expression (red arrow) across the Wanderlust trajectory taken in segments of 0.1 C) Distribution of marker expression across the trajectory for CD24, TdT, and CD10. D) Marker traces across the trajectory for four different samples (denoted a to d) aligned using cross-correlation. Pearson's ρ > 0.9 between the trajectories of different samples. The red box demarcates the expression of CD24, which bisects the TdT expression prior to CD10 expression across all four healthy individuals. See also Figure S2 for traces on full marker panel and additional robustness analysis.
Figure 3. Wanderlust uncovers rare B cell progenitors prior to the expression of CD10 or CD19
A) Wanderlust trace showing the expression of CD24, CD34, CD38, and TdT (upper left panel). Density plots of four distinct TdT and CD24 populations defined within the CD34+CD38+ fraction (right). A histogram overlay of the Wanderlust values for cells contained in population I-V (dark blue boxes). The median Wanderlust values are indicated. B) Expression of the surrogate light chain of the pre-B cell receptor, λ5 and VpreB (top), and CD10 and IgH (i)ntracellular (bottom) across populations I though V. ‘*” denotes the uniform expression of IgHi in population V. C) Relative to population II, the amount of (C) IgH DJ and (D) IgH V(D)J rearrangement by qPCR of genomic DNA from prospectively isolated cells from populations II-V. Triplicate analysis of two biological replicates. Error bars are standard deviation. ‘*’ denotes uniform IgHi expression implying complete IgH V(D)J rearrangement. E) Pie charts summarizing the cellular contribution of human BMMNCs. The purple pie denotes CD34+CD38+ cells. Percentages are relative to all BMMNCs. See also Figure S3 for supporting data.
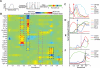
Figure 4. Regulatory signaling re-wires across development
A) Compared to basal control, the IL-7 induction of pSTAT5 in populations I through V. Population III (CD34+CD38+TdT+) had consistently the largest induction across seven replicate BM. Induction is arcsinh difference versus basal, scaled individually. Maximum differences were 0.8, 2.2, 1.7, 1.3, 0.3, 0.8, and 1.8 for A to G, respectively. B) The effect of TSLP, tofacitinib (JAK1/3i), and pervanadate (PVO) on induction of pSTAT5 across populations I to V in the same individual versus the basal. The maximum difference is 1.3. C) Early Wanderlust trace (trajectory 0-0.5) showing expression of IL-7rα (CD127), TdT, and IgHi. The green heat bar indicates relative pSTAT5 phosphorylation in the basal sample. Relative positions of populations I to V and IgH V(D)J rearrangement are indicated. D) Network rewiring between population III and IV. The schematics show proposed regulatory networks of pSTAT5; population III activates in a ligand and JAK-dependent manner whereas population IV becomes ligand independent, but maintains a high pSTAT5 level in the absence of stimulation. See also Figure S4 and table S2 for additional perturbations and full distributions representing the IL-7 perturbation.
Figure 5. Coordination of protein expression across B cell development
A) The first derivative was calculated in windows across the trajectory for each marker. These values are expressed as a heat map with red indicating a positive slope (increasing expression) and blue indicating a negative slope (decreasing expression). B) Heat map summary of the first derivative analysis, rows are markers and columns the progression of the trajectory. Markers were hierarchically clustered using absolute values of the first derivative. ‘*’ indicates decline in kappa light chain expression coincident with increase in lambda light chain expression. Coordination points: C) at ~0.25 Wanderlust (red dashed line and box) rise in VpreB, λ5, TdT, CD10, and CD24 and fall in CD117 expression; D) 0.3-0.4 (blue dashed line and box) drop in CD34 expression and increases in CD19, CD20, IgHi, and Pax5; E) ~0.6 (purple dashed line and box) showing increases in CD20, kappa, and lambda light chain protein expression. F) ~0.8 (black dashed line and box) with drop in CD38 expression and increases in CD40, IgD, and CD22. See also Figure S5 for additional replicates.
Figure 6. Regulatory signaling influences cell fate decisions in developing B cells
A) Wanderlust traces of VpreB, λ5, IgHi, and cleaved PARP across the early trajectory (0.1-0.6). The background is shaded for Ki67 (proliferative antigen) expression. Indicated are the relative positions of population II to V. The red and blue arrows indicate putative timing of pro- and pre-B cell coordination points, respectively. B) Wanderlust traces across the late trajectory (0.3-0.9) showing IgH (i)ntracellular and (s)urface expression with the basal and receptor cross-linked levels of pPLCϒ2, an intermediary of B cell receptor signaling. pPLCϒ2 induction in response to cross-link shown in yellow. C) Using two human BM samples, B cell differentiation co-cultures were performed, following lineage depletion, for 6 weeks on an OP-9 stromal layer in the presence or absence of inhibitors. After, cells were analyzed by flow cytometry for the frequencies of populations II through IV. D) Frequency of populations II through IV, as a proportion of CD34+CD38+ cells, after six weeks of culture in the indicated condition. Each treatment is normalized to the DMSO control for that population. Two biological replicates analyzed in quadruplicate cultures for each condition. Error bars are standard deviation. A two-tailed t-test was performed to determine statistical significance *p<0.05, **p<0.01, ***p<0.001. See also Figure S6 and Table S4 for supporting data.
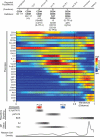
Figure 7. Summary of phenotypic marker expression, regulatory signaling, and IgH recombination status and proposed pro- and pre- B cell checkpoints aligned to reported populations across human B cell development as ordered by the Wanderlust trajectory
Under ‘Definition’ (top) the expected marker localization and expression is listed for subsequent cytometric characterization. ‘Bold type’ markers are the minimum required to positively select cells by cytometric analysis. ‘Regular type’ markers are uniformly expressed on the surface. ‘Italic type’ markers are intracellularly expressed. Markers in parentheses ‘()’ are only partially expressed on that fraction. Red and blue bars indicate the pro- and pre- B cell coordination points, respectively. For regulatory signaling and IgH status (bottom), the grey bars indicate the developmental regions where cells are prone to the listed behaviors. See also Figure S7 for additional replicates.
Similar articles
- Hind limb unloading, a model of spaceflight conditions, leads to decreased B lymphopoiesis similar to aging.
Lescale C, Schenten V, Djeghloul D, Bennabi M, Gaignier F, Vandamme K, Strazielle C, Kuzniak I, Petite H, Dosquet C, Frippiat JP, Goodhardt M. Lescale C, et al. FASEB J. 2015 Feb;29(2):455-63. doi: 10.1096/fj.14-259770. Epub 2014 Nov 5. FASEB J. 2015. PMID: 25376832 - Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development.
Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Malin S, et al. Nat Immunol. 2010 Feb;11(2):171-9. doi: 10.1038/ni.1827. Epub 2009 Nov 29. Nat Immunol. 2010. PMID: 19946273 Free PMC article. - De novo DNA Methyltransferases Dnmt3a and Dnmt3b regulate the onset of Igκ light chain rearrangement during early B-cell development.
Manoharan A, Du Roure C, Rolink AG, Matthias P. Manoharan A, et al. Eur J Immunol. 2015 Aug;45(8):2343-55. doi: 10.1002/eji.201445035. Epub 2015 Jun 16. Eur J Immunol. 2015. PMID: 26059604 - Fate decisions regulating bone marrow and peripheral B lymphocyte development.
Monroe JG, Dorshkind K. Monroe JG, et al. Adv Immunol. 2007;95:1-50. doi: 10.1016/S0065-2776(07)95001-4. Adv Immunol. 2007. PMID: 17869609 Review. - The key role of IL-7 in lymphopoiesis.
Ceredig R, Rolink AG. Ceredig R, et al. Semin Immunol. 2012 Jun;24(3):159-64. doi: 10.1016/j.smim.2012.02.004. Epub 2012 Mar 14. Semin Immunol. 2012. PMID: 22421573 Review.
Cited by
- Central human B cell tolerance manifests with a distinctive cell phenotype and is enforced via CXCR4 signaling in hu-mice.
Alves da Costa T, Peterson JN, Lang J, Shulman J, Liang X, Freed BM, Boackle SA, Lauzurica P, Torres RM, Pelanda R. Alves da Costa T, et al. Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2021570118. doi: 10.1073/pnas.2021570118. Proc Natl Acad Sci U S A. 2021. PMID: 33850015 Free PMC article. - The Shoot Apical Meristem: An Evolutionary Molding of Higher Plants.
Kean-Galeno T, Lopez-Arredondo D, Herrera-Estrella L. Kean-Galeno T, et al. Int J Mol Sci. 2024 Jan 26;25(3):1519. doi: 10.3390/ijms25031519. Int J Mol Sci. 2024. PMID: 38338798 Free PMC article. Review. - Computational flow cytometry: helping to make sense of high-dimensional immunology data.
Saeys Y, Van Gassen S, Lambrecht BN. Saeys Y, et al. Nat Rev Immunol. 2016 Jul;16(7):449-62. doi: 10.1038/nri.2016.56. Epub 2016 Jun 20. Nat Rev Immunol. 2016. PMID: 27320317 Review. - A new experimental platform facilitates assessment of the transcriptional and chromatin landscapes of aging yeast.
Hendrickson DG, Soifer I, Wranik BJ, Kim G, Robles M, Gibney PA, McIsaac RS. Hendrickson DG, et al. Elife. 2018 Oct 19;7:e39911. doi: 10.7554/eLife.39911. Elife. 2018. PMID: 30334737 Free PMC article. - Branch-recombinant Gaussian processes for analysis of perturbations in biological time series.
Penfold CA, Sybirna A, Reid JE, Huang Y, Wernisch L, Ghahramani Z, Grant M, Surani MA. Penfold CA, et al. Bioinformatics. 2018 Sep 1;34(17):i1005-i1013. doi: 10.1093/bioinformatics/bty603. Bioinformatics. 2018. PMID: 30423108 Free PMC article.
References
- Cobaleda C, Sanchez-García I. B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. BioEssays. 2009;31:600–609. - PubMed
- Corfe SA, Paige CJ. The many roles of IL-7 in B cell development; Mediator of survival, proliferation and differentiation. Semin Immunol. 2012;24:198–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA164729/CA/NCI NIH HHS/United States
- HHSF223201210194C/PHS HHS/United States
- U54CA121852-01A1/CA/NCI NIH HHS/United States
- N01-HV-00242/HV/NHLBI NIH HHS/United States
- 41000411217/PHS HHS/United States
- U54CA149145/CA/NCI NIH HHS/United States
- 152175.5041015.0412/PHS HHS/United States
- UL1 RR025744/RR/NCRR NIH HHS/United States
- UL1 TR001085/TR/NCATS NIH HHS/United States
- 5U54CA143907/CA/NCI NIH HHS/United States
- U54 CA149145/CA/NCI NIH HHS/United States
- HHSN272200700038C/AI/NIAID NIH HHS/United States
- DP2-OD002414-01/OD/NIH HHS/United States
- U24 CA224309/CA/NCI NIH HHS/United States
- 5-24927/PHS HHS/United States
- R01 CA130826/CA/NCI NIH HHS/United States
- P01 CA034233-22A1/CA/NCI NIH HHS/United States
- U54 CA143907/CA/NCI NIH HHS/United States
- PN2 EY018228/EY/NEI NIH HHS/United States
- S10 SIG S1044027582-01/PHS HHS/United States
- P01 CA034233/CA/NCI NIH HHS/United States
- K99GM104148-01/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- DP2 OD002414/OD/NIH HHS/United States
- S10 RR027582/RR/NCRR NIH HHS/United States
- U19 AI100627/AI/NIAID NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- 1R01CA130826/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- K99 GM104148/GM/NIGMS NIH HHS/United States
- R00 GM104148/GM/NIGMS NIH HHS/United States
- PN2EY018228/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous