Natural selection and infectious disease in human populations - PubMed (original) (raw)
Review
Natural selection and infectious disease in human populations
Elinor K Karlsson et al. Nat Rev Genet. 2014 Jun.
Abstract
The ancient biological 'arms race' between microbial pathogens and humans has shaped genetic variation in modern populations, and this has important implications for the growing field of medical genomics. As humans migrated throughout the world, populations encountered distinct pathogens, and natural selection increased the prevalence of alleles that are advantageous in the new ecosystems in both host and pathogens. This ancient history now influences human infectious disease susceptibility and microbiome homeostasis, and contributes to common diseases that show geographical disparities, such as autoimmune and metabolic disorders. Using new high-throughput technologies, analytical methods and expanding public data resources, the investigation of natural selection is leading to new insights into the function and dysfunction of human biology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
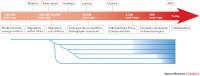
Figure 1. Pathogen emergence during human history.
Key events in recent human evolution (boxes outlined in black) are juxtaposed with the estimated ages of infectious disease emergence (boxes outlined in red). The fragmentation of the human lineage into genetically and geographically distinct populations (blue lines) accelerates with migration out of Africa. Later, these populations started mixing more (blue shaded regions between the populations) along trade routes (such as the Silk Road), through colonization and through high rates of global travel nowadays. PowerPoint slide
Figure 2. Positive selection increases power to detect associations in GWASs.
a | The variants in a population-wide sample are shown for a schematic genomic locus. The red region indicates a variant that provides selective advantage (such as a host variant that confers relative resistance to an infectious agent). Positive selection of that variant rapidly increases its prevalence in a population and also the prevalence of nearby alleles that are in linkage with it. b | Positive selection on a variant is detectable with three types of signals: high levels of differentiation (that is, when positive selection in one geographical region causes larger frequency differences between populations than those expected for neutrally evolving alleles); high frequency of the derived allele (that is, when a new allele increases to a frequency higher than that expected under genetic drift); and long haplotypes (as determined by the integrated haplotype score and cross-population extended haplotype homozygosity) that are left when the selected allele increases in frequency sufficiently quickly that long-range associations with neighbouring variants are maintained. Combining different signals of selection into a composite score can increase resolution by up to 100-fold, which facilitates identification of the causal variants. c | Variants that are of high frequency, of strong effects and in regions of extensive linkage disequilibrium (LD) — all of which are characteristics of positively selected loci — are detectable with smaller sample sizes in genome-wide association studies (GWASs). Even with full sequence data, low-frequency alleles (solid blue lines) require larger sample sizes (x axis; equal number of cases and controls) than high-frequency alleles (solid red lines) for equivalent power over a range of effect sizes (1.2, 1.5 and 2.0). When mapping with genotyping arrays, shorter LD in unselected regions (blue dashed lines; modelled here using power simulations for the sparser Illumina 300K array) can cause a larger loss of power (relative to full sequence data) than in selected regions with longer LD (red dashed lines; modelled as denser Illumina 1M array). _F_ST, Wright's fixation index. Part c is adapted from Ref. . PowerPoint slide
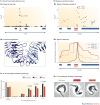
Figure 3. Selected variants implicated in pathogen resistance.
A | A genome-wide scan for signals of positive selection in the Yoruban population of Nigeria found a strongly selected nonsynonymous single-nucleotide polymorphism (SNP) that alters the pathogen recognition protein Toll-like receptor 5 (TLR5) (part Aa) and that is predicted to disrupt TLR5 activation in response to flagellated bacteria (part Ab). Cell lines that carry the new TLR5 variant (Leu616Phe) had significantly reduced nuclear factor-κB (NF-κB) signalling in response to flagellin, which is potentially protective against some bacterial infections (part Ac). Error bars represent the standard error of the mean over at least three independent experiments; P values are indicated above the bar graphs. B | Two common variants of the apolipoprotein L1 (APOL1) gene (Allele G1 and Allele G2) that are strongly associated with kidney disease in African Americans (part Ba) show evidence of recent positive selection in Yorubans (part Bb). In vitro, the G1 and G2 variants lyse subspecies of the Trypanosoma spp. pathogen that are resistant to wild-type APOL1 (part Bc). Arrows point to the swelling lysosome. cM, centimorgan; PMA, phorbol myristate acetate. Part A reprinted from Cell, 152, Sharon R. Grossman et al., Identifying recent adaptations in large-scale genomic data, 703–713, © (2013), with permission from Elsevier. Part B from Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010). Reprinted with permission from AAAS. PowerPoint slide
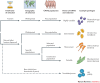
Figure 4. The power offered by combining natural selection with GWASs depends on the age of selection and populations chosen.
For pathogens that predate human dispersal from Africa, ancient and complex signals of selection are shared between human populations, and there are variable implications for genome-wide association studies (GWASs). For widespread pathogens that are more recent, the range of new resistance variants will be more limited, but selection is harder to detect when it is shared between populations. For recent pathogens that affect specific populations, GWASs in the selected populations will be particularly powerful, as causal variants will have been driven to high prevalence. Selection signals from one population may help to detect resistance loci in other unselected populations, but only if resistance variants arise in the same genetic loci. For GWASs of very new diseases, supplementing these studies with methods to detect selection will add no power, unless variants that confer resistance also protect against more ancient pathogens. Examples of pathogens matching each scenario are given on the right. PowerPoint slide
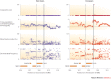
Figure 5. Signals of selection and association may differ between populations.
Two loci (represented by dashed and solid lines) associated with leprosy susceptibility in Han Chinese, show no evidence of positive selection in East Asians (blue circles). The association downstream of the cylindromatosis (CYLD) gene (solid line), but not the association near the bacterial pattern recognition receptor gene NOD2 (dashed line), has signals of positive selection in Europeans (red circles). Selection scores for four different metrics were calculated using data from the 1000 Genomes Project and published in Ref. . _F_ST, Wright's fixation index; NKD1, naked cuticle homologue 1; SNX20, sorting nexin 20. PowerPoint slide
Similar articles
- Emerging prion disease drives host selection in a wildlife population.
Robinson SJ, Samuel MD, Johnson CJ, Adams M, McKenzie DI. Robinson SJ, et al. Ecol Appl. 2012 Apr;22(3):1050-9. doi: 10.1890/11-0907.1. Ecol Appl. 2012. PMID: 22645831 - Evolution of cytokine production capacity in ancient and modern European populations.
Domínguez-Andrés J, Kuijpers Y, Bakker OB, Jaeger M, Xu CJ, Van der Meer JW, Jakobsson M, Bertranpetit J, Joosten LA, Li Y, Netea MG. Domínguez-Andrés J, et al. Elife. 2021 Sep 7;10:e64971. doi: 10.7554/eLife.64971. Elife. 2021. PMID: 34488939 Free PMC article. - Systematic detection of positive selection in the human-pathogen interactome and lasting effects on infectious disease susceptibility.
Corona E, Wang L, Ko D, Patel CJ. Corona E, et al. PLoS One. 2018 May 25;13(5):e0196676. doi: 10.1371/journal.pone.0196676. eCollection 2018. PLoS One. 2018. PMID: 29799843 Free PMC article. - The impact of "ancient pathogen" studies on the practice of public health.
Greenblatt C, Spigelman M, Vernon K. Greenblatt C, et al. Public Health Rev. 2003;31(2):81-91. Public Health Rev. 2003. PMID: 15255158 Review. - Infectious disease in the genomic era.
Yang X, Yang H, Zhou G, Zhao GP. Yang X, et al. Annu Rev Genomics Hum Genet. 2008;9:21-48. doi: 10.1146/annurev.genom.9.081307.164428. Annu Rev Genomics Hum Genet. 2008. PMID: 18767959 Review.
Cited by
- Signals of positive selection in genomes of palearctic Myotis-bats coexisting with a fungal pathogen.
Twort VG, Laine VN, Field KA, Whiting-Fawcett F, Ito F, Reiman M, Bartonicka T, Fritze M, Ilyukha VA, Belkin VV, Khizhkin EA, Reeder DM, Fukui D, Jiang TL, Lilley TM. Twort VG, et al. BMC Genomics. 2024 Sep 3;25(1):828. doi: 10.1186/s12864-024-10722-3. BMC Genomics. 2024. PMID: 39227786 Free PMC article. - Acute soft head syndrome in a teenager with sickle cell anemia: A case report.
Magitta NF, Komanya FB, Alphonce BO, Bitesigilwe MD, Sindato EM, Meda JR. Magitta NF, et al. Clin Case Rep. 2023 Nov 6;11(11):e8174. doi: 10.1002/ccr3.8174. eCollection 2023 Nov. Clin Case Rep. 2023. PMID: 37942183 Free PMC article. - Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae.
Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, Mawejje HD, Djogbenou LS, Steen K, Rippon EJ, Clarkson CS, Field SG, Rigden DJ, Donnelly MJ. Weetman D, et al. Mol Ecol. 2015 Jun;24(11):2656-72. doi: 10.1111/mec.13197. Epub 2015 May 14. Mol Ecol. 2015. PMID: 25865270 Free PMC article. - Role of genetic factors and ethnicity on the multiplicity of Plasmodium falciparum infection in children with asymptomatic malaria in Yaoundé, Cameroon.
Roman DNR, Anne NNR, Singh V, Luther KMM, Chantal NEM, Albert MS. Roman DNR, et al. Heliyon. 2018 Aug 30;4(8):e00760. doi: 10.1016/j.heliyon.2018.e00760. eCollection 2018 Aug. Heliyon. 2018. PMID: 30186982 Free PMC article. - Admixture-enabled selection for rapid adaptive evolution in the Americas.
Norris ET, Rishishwar L, Chande AT, Conley AB, Ye K, Valderrama-Aguirre A, Jordan IK. Norris ET, et al. Genome Biol. 2020 Feb 7;21(1):29. doi: 10.1186/s13059-020-1946-2. Genome Biol. 2020. PMID: 32028992 Free PMC article.
References
- Polgar S, Tax S. Horizons of Anthropology. 1964.
- Armelagos GJ, Barnes KC, Lin J. Disease in human evolution: the re-emergence of infectious disease in the third epidemiological transition. AnthroNotes. 1996;18:1–7. doi: 10.5479/10088/22354. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials