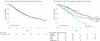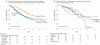Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs - PubMed (original) (raw)
Clinical Trial
. 2014 May 21;311(19):1998-2006.
doi: 10.1001/jama.2014.3741.
Bruce E Johnson 2, Lynne D Berry 3, David J Kwiatkowski 4, A John Iafrate 5, Ignacio I Wistuba 6, Marileila Varella-Garcia 7, Wilbur A Franklin 7, Samuel L Aronson 8, Pei-Fang Su 3, Yu Shyr 3, D Ross Camidge 7, Lecia V Sequist 5, Bonnie S Glisson 6, Fadlo R Khuri 9, Edward B Garon 10, William Pao 3, Charles Rudin 11, Joan Schiller 12, Eric B Haura 13, Mark Socinski 14, Keisuke Shirai 15, Heidi Chen 3, Giuseppe Giaccone 16, Marc Ladanyi 1, Kelly Kugler 7, John D Minna 12, Paul A Bunn 7
Affiliations
- PMID: 24846037
- PMCID: PMC4163053
- DOI: 10.1001/jama.2014.3741
Clinical Trial
Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs
Mark G Kris et al. JAMA. 2014.
Abstract
Importance: Targeting oncogenic drivers (genomic alterations critical to cancer development and maintenance) has transformed the care of patients with lung adenocarcinomas. The Lung Cancer Mutation Consortium was formed to perform multiplexed assays testing adenocarcinomas of the lung for drivers in 10 genes to enable clinicians to select targeted treatments and enroll patients into clinical trials.
Objectives: To determine the frequency of oncogenic drivers in patients with lung adenocarcinomas and to use the data to select treatments targeting the identified driver(s) and measure survival.
Design, setting, and participants: From 2009 through 2012, 14 sites in the United States enrolled patients with metastatic lung adenocarcinomas and a performance status of 0 through 2 and tested their tumors for 10 drivers. Information was collected on patients, therapies, and survival.
Interventions: Tumors were tested for 10 oncogenic drivers, and results were used to select matched targeted therapies.
Main outcomes and measures: Determination of the frequency of oncogenic drivers, the proportion of patients treated with genotype-directed therapy, and survival.
Results: From 2009 through 2012, tumors from 1007 patients were tested for at least 1 gene and 733 for 10 genes (patients with full genotyping). An oncogenic driver was found in 466 of 733 patients (64%). Among these 733 tumors, 182 tumors (25%) had the KRAS driver; sensitizing EGFR, 122 (17%); ALK rearrangements, 57 (8%); other EGFR, 29 (4%); 2 or more genes, 24 (3%); ERBB2 (formerly HER2), 19 (3%); BRAF, 16 (2%); PIK3CA, 6 (<1%); MET amplification, 5 (<1%); NRAS, 5 (<1%); MEK1, 1 (<1%); AKT1, 0. Results were used to select a targeted therapy or trial in 275 of 1007 patients (28%). The median survival was 3.5 years (interquartile range [IQR], 1.96-7.70) for the 260 patients with an oncogenic driver and genotype-directed therapy compared with 2.4 years (IQR, 0.88-6.20) for the 318 patients with any oncogenic driver(s) who did not receive genotype-directed therapy (propensity score-adjusted hazard ratio, 0.69 [95% CI, 0.53-0.9], P = .006).
Conclusions and relevance: Actionable drivers were detected in 64% of lung adenocarcinomas. Multiplexed testing aided physicians in selecting therapies. Although individuals with drivers receiving a matched targeted agent lived longer, randomized trials are required to determine if targeting therapy based on oncogenic drivers improves survival.
Trial registration: clinicaltrials.gov Identifier: NCT01014286.
Figures
Figure 1. Survival of Patients
ALK indicates anaplastic lymphoma kinase gene; EGFR(s), epidermal growth factor receptor gene (sensitizing); EGFR(o), epidermal growth factor receptor gene (other); KRAS, Kirsten rat sarcoma; NA, not applicable. A, The median survival 2.65 years (95% CI, 2.35-2.93). B, Median survival (95% CI): EGFR(s), 3.97 years (3.21-4.64); EGFR(o), 2.70 years (1.42-NA); ALK, 4.25 years (2.92-NA); KRAS, 2.41 years (1.87-3.21); doubletons (oncogenic drivers in 2 genes), 2.03 years (1.39-2.84). Vertical tick marks are censoring events.
Figure 2. Survival Comparisons
ALK indicates anaplastic lymphoma kinase gene; EGFR(s), epidermal growth factor receptor gene (sensitizing); EGFR(o), epidermal growth factor receptor gene (other); KRAS, Kirsten rat sarcoma; NA, not applicable. A, Median survival (95% CI): oncogenic driver + no targeted therapy, 2.38 (1.81-2.93); oncogenic driver + targeted therapy, 3.49 (3.02-4.33); no oncogenic driver, 2.08 (1.84-2.46). B, Survival by oncogenic driver detected for patients with the 5 most frequent oncogenic drivers and targeted treatment. Median survival (95% CI): EGFR(s), 3.78 (2.77-NA); EGFR(o), 2.70 (1.42-NA); ALK, NA (2.80-NA); KRAS, 4.85 (1.30-NA); doubletons (oncogenic drivers in 2 genes), 2.69 (1.94-NA). Vertical tick marks are censoring events.
Comment in
- Non-small cell lung cancer and precision medicine: a model for the incorporation of genomic features into clinical trial design.
Pasche B, Grant SC. Pasche B, et al. JAMA. 2014 May 21;311(19):1975-6. doi: 10.1001/jama.2014.3742. JAMA. 2014. PMID: 24846033 No abstract available.
Similar articles
- Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: Results from the Lung Cancer Mutation Consortium.
Steuer CE, Behera M, Berry L, Kim S, Rossi M, Sica G, Owonikoko TK, Johnson BE, Kris MG, Bunn PA, Khuri FR, Garon EB, Ramalingam SS. Steuer CE, et al. Cancer. 2016 Mar 1;122(5):766-72. doi: 10.1002/cncr.29812. Epub 2015 Dec 22. Cancer. 2016. PMID: 26695526 Free PMC article. - [Oncogenic drivers in daily practice improve overall survival in patients with lung adenocarcinoma].
Fournier C, Greillier L, Fina F, Secq V, Nanni-Metellus I, Loundou A, Garcia S, Ouafik L, Tomasini P, Barlesi F. Fournier C, et al. Rev Mal Respir. 2016 Nov;33(9):751-756. doi: 10.1016/j.rmr.2015.12.009. Epub 2016 Mar 24. Rev Mal Respir. 2016. PMID: 27017063 French. - Clinicopathologic features and outcomes of patients with lung adenocarcinomas harboring BRAF mutations in the Lung Cancer Mutation Consortium.
Villaruz LC, Socinski MA, Abberbock S, Berry LD, Johnson BE, Kwiatkowski DJ, Iafrate AJ, Varella-Garcia M, Franklin WA, Camidge DR, Sequist LV, Haura EB, Ladanyi M, Kurland BF, Kugler K, Minna JD, Bunn PA, Kris MG. Villaruz LC, et al. Cancer. 2015 Feb 1;121(3):448-56. doi: 10.1002/cncr.29042. Epub 2014 Oct 1. Cancer. 2015. PMID: 25273224 Free PMC article. - Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches.
Shea M, Costa DB, Rangachari D. Shea M, et al. Ther Adv Respir Dis. 2016 Apr;10(2):113-29. doi: 10.1177/1753465815617871. Epub 2015 Nov 30. Ther Adv Respir Dis. 2016. PMID: 26620497 Free PMC article. Review. - Successes and limitations of targeted cancer therapy in lung cancer.
Suda K, Mitsudomi T. Suda K, et al. Prog Tumor Res. 2014;41:62-77. doi: 10.1159/000355902. Epub 2014 Feb 17. Prog Tumor Res. 2014. PMID: 24727987 Review.
Cited by
- Treatment Patterns and Survival of Patients With Advanced Non-Small Cell Lung Cancer Guided by Comprehensive Genomic Profiling: Real-World Single-Institute Study in China.
Lv W, Cheng H, Shao D, Wei Y, Zhu W, Wu K, Jiang W, Hu L, Sha Z, Zhong B, Pei X. Lv W, et al. Front Oncol. 2021 Mar 10;11:630717. doi: 10.3389/fonc.2021.630717. eCollection 2021. Front Oncol. 2021. PMID: 33777783 Free PMC article. - Value: The Next Frontier in Cancer Care.
Goulart BH. Goulart BH. Oncologist. 2016 Jun;21(6):651-3. doi: 10.1634/theoncologist.2016-0174. Epub 2016 May 25. Oncologist. 2016. PMID: 27226360 Free PMC article. - Testing for ROS1 in non-small cell lung cancer: a review with recommendations.
Bubendorf L, Büttner R, Al-Dayel F, Dietel M, Elmberger G, Kerr K, López-Ríos F, Marchetti A, Öz B, Pauwels P, Penault-Llorca F, Rossi G, Ryška A, Thunnissen E. Bubendorf L, et al. Virchows Arch. 2016 Nov;469(5):489-503. doi: 10.1007/s00428-016-2000-3. Epub 2016 Aug 17. Virchows Arch. 2016. PMID: 27535289 Free PMC article. Review. - Referred molecular testing as a barrier to optimal treatment decision making in metastatic non-small cell lung cancer: Experience at a tertiary academic institution in Canada.
Grafham GK, Craddock KJ, Huang WY, Louie AV, Zhang L, Hwang DM, Parmar A. Grafham GK, et al. Cancer Med. 2024 Feb;13(3):e6886. doi: 10.1002/cam4.6886. Epub 2024 Feb 5. Cancer Med. 2024. PMID: 38317584 Free PMC article. - What does radiomics do in PD-L1 blockade therapy of NSCLC patients?
Cui R, Yang Z, Liu L. Cui R, et al. Thorac Cancer. 2022 Oct;13(19):2669-2680. doi: 10.1111/1759-7714.14620. Epub 2022 Aug 29. Thorac Cancer. 2022. PMID: 36039482 Free PMC article. Review.
References
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. - PubMed
- Pao W, Girard N. New driver mutations in non-small cell lung cancer. Lancet Oncol. 2011;12(2):175–180. - PubMed
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. - PubMed
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- P50 CA058187/CA/NCI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- P01 CA129243/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- 1RC2CA148394-010/CA/NCI NIH HHS/United States
- K23 CA149079/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous