Protein flexibility facilitates quaternary structure assembly and evolution - PubMed (original) (raw)
Protein flexibility facilitates quaternary structure assembly and evolution
Joseph A Marsh et al. PLoS Biol. 2014.
Abstract
The intrinsic flexibility of proteins allows them to undergo large conformational fluctuations in solution or upon interaction with other molecules. Proteins also commonly assemble into complexes with diverse quaternary structure arrangements. Here we investigate how the flexibility of individual protein chains influences the assembly and evolution of protein complexes. We find that flexibility appears to be particularly conducive to the formation of heterologous (i.e., asymmetric) intersubunit interfaces. This leads to a strong association between subunit flexibility and homomeric complexes with cyclic and asymmetric quaternary structure topologies. Similarly, we also observe that the more nonhomologous subunits that assemble together within a complex, the more flexible those subunits tend to be. Importantly, these findings suggest that subunit flexibility should be closely related to the evolutionary history of a complex. We confirm this by showing that evolutionarily more recent subunits are generally more flexible than evolutionarily older subunits. Finally, we investigate the very different explorations of quaternary structure space that have occurred in different evolutionary lineages. In particular, the increased flexibility of eukaryotic proteins appears to enable the assembly of heteromeric complexes with more unique components.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
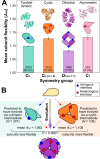
Figure 1. Relating the flexibility of homomeric subunits to quaternary structure topology and evolution.
(A) Comparison of subunit flexibility, as measured by A rel, for homomers from different symmetry groups. An example from each symmetry group is shown above. The numbers and percentages of each group within the total set of homomeric complexes are shown on the bars. These groups comprise all complexes in the PDB except the rare cubic (0.9%) and helical (0.6%) symmetry groups. Error bars represent SEM. Boxplots for each group along with the p values between groups are provided in Figure S2A. (B) There are two possible evolutionary pathways for a dihedral hexamer (D 3): via a twofold dimer (C 2) intermediate (left) or via a cyclic (C 3) intermediate (right). When considering all such complexes where two different evolutionary pathways are possible, we observe a strong tendency for those that evolved via a cyclic intermediate to have more flexible subunits. Interestingly, the subunits of complexes with predicted dimeric intermediates are less flexible than those from twofold dimeric complexes (A rel = 1.063 versus 1.099, p = 5×10−7, Wilcoxon rank-sum test) and those from complexes with predicted cyclic intermediates are less flexible (but not significantly so) than those from cyclic complexes (A rel = 1.108 versus 1.127, p = 0.7). One potential explanation for this is that lower subunit flexibility might be associated with a greater propensity for evolving higher order quaternary structures via dimeric or cyclic intermediates.
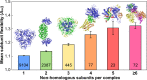
Figure 2. Comparison of subunit flexibility from protein complexes with varying numbers of nonhomologous subunits.
Examples of complexes with varying numbers of nonhomologous subunits are shown above. The numbers of unique chains in each group are shown on the bars. Error bars represent SEM. Boxplots for each group are provided in Figure S2B.
Figure 3. The importance of protein flexibility for the evolution of new heteromeric subunits.
(A) Model for the evolution of heteromeric complexes in which new subunits of increasing flexibility are sequentially gained. (B) Example of a protein complex (Gβ5-RGS9, PDB ID: 2PBI) in which different relative ages can be assigned to different subunits. There is an ortholog of Gβ5 (blue) in yeast, whereas no orthologs or domain-architecture homologs of RGS9 (yellow) can be detected in yeast or any other species of a similar or greater evolutionary distance from humans (the most distant ortholog is observed in Caenorhabditis elegans and the most distant homolog sharing the same domain architecture is seen in the single-celled eukaryote Capsaspora owczarzaki, which is more closely related to humans than yeast). (C) Pairwise comparisons of the flexibility of putative older and putative newer subunits of human (or closely related) protein complexes, with respect to different species. No species more closely related to humans than C. elegans and Drosophila melanogaster are shown as there are none where >5 complexes with putative older and newer subunits can be identified. The full set of species considered is provided in Table S2. The p values calculated with the Wilcoxon signed-rank test are shown for each species, and the numbers of complexes from each species are shown in parentheses. Error bars represent SEM.
Figure 4. The relationship between evolution, quaternary structure topology, and protein flexibility.
(A) Comparison of the numbers of homomers, homologous heteromers (i.e., heteromers where all distinct chains are homologs), and nonhomologous heteromers from bacteria, archaea, and eukaryotes. (B) Comparison of the mean subunit flexibility and number of nonhomologous subunits per complex for the 50 species with the most complexes in our dataset. Values for all species are provided in Table S3. (C) Comparison of subunit flexibility for protein complexes with varying numbers of nonhomologous subunits from bacteria, archaea, and eukaryotes. Error bars represent SEM. A similar species-level analysis is provided in Figure S8.
Similar articles
- Structure, dynamics, assembly, and evolution of protein complexes.
Marsh JA, Teichmann SA. Marsh JA, et al. Annu Rev Biochem. 2015;84:551-75. doi: 10.1146/annurev-biochem-060614-034142. Epub 2014 Dec 8. Annu Rev Biochem. 2015. PMID: 25494300 Review. - Coevolutionary constraints in the sequence-space of macromolecular complexes reflect their self-assembly pathways.
Mallik S, Kundu S. Mallik S, et al. Proteins. 2017 Jul;85(7):1183-1189. doi: 10.1002/prot.25292. Epub 2017 Apr 6. Proteins. 2017. PMID: 28342228 - Comparative analyses of quaternary arrangements in homo-oligomeric proteins in superfamilies: Functional implications.
Sudha G, Srinivasan N. Sudha G, et al. Proteins. 2016 Sep;84(9):1190-202. doi: 10.1002/prot.25065. Epub 2016 Jun 8. Proteins. 2016. PMID: 27177429 - Parallel dynamics and evolution: Protein conformational fluctuations and assembly reflect evolutionary changes in sequence and structure.
Marsh JA, Teichmann SA. Marsh JA, et al. Bioessays. 2014 Feb;36(2):209-18. doi: 10.1002/bies.201300134. Epub 2013 Nov 25. Bioessays. 2014. PMID: 24272815 Review. - Structural, evolutionary, and assembly principles of protein oligomerization.
Levy ED, Teichmann S. Levy ED, et al. Prog Mol Biol Transl Sci. 2013;117:25-51. doi: 10.1016/B978-0-12-386931-9.00002-7. Prog Mol Biol Transl Sci. 2013. PMID: 23663964 Review.
Cited by
- Structural and mechanistic insights into human splicing factor SF3b complex derived using an integrated approach guided by the cryo-EM density maps.
Rakesh R, Joseph AP, Bhaskara RM, Srinivasan N. Rakesh R, et al. RNA Biol. 2016 Oct 2;13(10):1025-1040. doi: 10.1080/15476286.2016.1218590. Epub 2016 Aug 11. RNA Biol. 2016. PMID: 27618338 Free PMC article. - Unravelling the molecular interactions between the SARS-CoV-2 RBD spike protein and various specific monoclonal antibodies.
Martí D, Alsina M, Alemán C, Bertran O, Turon P, Torras J. Martí D, et al. Biochimie. 2022 Feb;193:90-102. doi: 10.1016/j.biochi.2021.10.013. Epub 2021 Oct 25. Biochimie. 2022. PMID: 34710552 Free PMC article. - Phosphorylation of the Bacillus subtilis Replication Controller YabA Plays a Role in Regulation of Sporulation and Biofilm Formation.
García García T, Ventroux M, Derouiche A, Bidnenko V, Correia Santos S, Henry C, Mijakovic I, Noirot-Gros MF, Poncet S. García García T, et al. Front Microbiol. 2018 Mar 21;9:486. doi: 10.3389/fmicb.2018.00486. eCollection 2018. Front Microbiol. 2018. PMID: 29619013 Free PMC article. - Reconstructing Ancient Proteins to Understand the Causes of Structure and Function.
Hochberg GKA, Thornton JW. Hochberg GKA, et al. Annu Rev Biophys. 2017 May 22;46:247-269. doi: 10.1146/annurev-biophys-070816-033631. Epub 2017 Mar 15. Annu Rev Biophys. 2017. PMID: 28301769 Free PMC article. Review. - Molecular ensembles make evolution unpredictable.
Sailer ZR, Harms MJ. Sailer ZR, et al. Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):11938-11943. doi: 10.1073/pnas.1711927114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078365 Free PMC article.
References
- Monod J, Wyman J, Changeux J-P (1965) On the nature of allosteric transitions: a plausible model. J Mol Biol 12: 88–118. - PubMed
- Perica T, Marsh JA, Sousa FL, Natan E, Colwell LJ, et al. (2012) The emergence of protein complexes: quaternary structure, dynamics and allostery. Biochem Soc Trans 40: 475–491. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources