DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin - PubMed (original) (raw)
DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin
Marina K Ayrapetov et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Dynamic changes in histone modification are critical for regulating DNA double-strand break (DSB) repair. Activation of the Tip60 acetyltransferase by DSBs requires interaction of Tip60 with histone H3 methylated on lysine 9 (H3K9me3). However, how H3K9 methylation is regulated during DSB repair is not known. Here, we demonstrate that a complex containing kap-1, HP1, and the H3K9 methyltransferase suv39h1 is rapidly loaded onto the chromatin at DSBs. Suv39h1 methylates H3K9, facilitating loading of additional kap-1/HP1/suv39h1 through binding of HP1's chromodomain to the nascent H3K9me3. This process initiates cycles of kap-1/HP1/suv39h1 loading and H3K9 methylation that facilitate spreading of H3K9me3 and kap-1/HP1/suv39h1 complexes for tens of kilobases away from the DSB. These domains of H3K9me3 function to activate the Tip60 acetyltransferase, allowing Tip60 to acetylate both ataxia telangiectasia-mutated (ATM) kinase and histone H4. Consequently, cells lacking suv39h1 display defective activation of Tip60 and ATM, decreased DSB repair, and increased radiosensitivity. Importantly, activated ATM rapidly phosphorylates kap-1, leading to release of the repressive kap-1/HP1/suv39h1 complex from the chromatin. ATM activation therefore functions as a negative feedback loop to remove repressive suv39h1 complexes at DSBs, which may limit DSB repair. Recruitment of kap-1/HP1/suv39h1 to DSBs therefore provides a mechanism for transiently increasing the levels of H3K9me3 in open chromatin domains that lack H3K9me3 and thereby promoting efficient activation of Tip60 and ATM in these regions. Further, transient formation of repressive chromatin may be critical for stabilizing the damaged chromatin and for remodeling the chromatin to create an efficient template for the DNA repair machinery.
Keywords: histone methylation; homologous recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
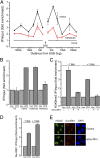
Fig. 1.
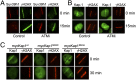
Suv39h1 promotes H3K9 methylation at DSBs. (A) 293T cells transfected with p84-ZFN were processed for ChIP by using γH2AX or H3K9me3 antibodies, followed by quantitative RT-PCR (RT-qPCR) with primer pairs located at the indicated positions. Results are fold enrichment relative to uncut DNA, which is assigned a value of 1 (solid black line; Uncut). Results are ±SD (n = 3). (B) 293T cells transfected with p84-ZFN (ZFN) or vector (Vec) were processed for ChIP by using antibodies to H3, γH2AX, H3K9me3, or H3K9me2 and primer pairs located 1.5 kb to the right of the DSB. Results are expressed as fold enrichment relative to uncut DNA. Results are ±SD (n = 3). (C) 293T cells were transfected with nonspecific (−) or siRNA targeting suv39h1 (+). Forty-eight hours later, cells were transfected with vector (Vec) or p84-ZFN (ZFN) and processed for ChIP by using antibody against H3K9me3 and primers located 1.5 kb to either side of the DSB. Results are ±SD (n = 3). (D) 293T cells were transfected with vector (Vec) or p84-ZFN (ZFN) and processed for ChIP with suv39h1 antibody and primers located 1.5 kb to either side of the DSB. Results are ±SD (n = 3). (E) U2OS cells were transfected with nonspecific (control) or suv39h1-specific (siSuv39h1) siRNA. Forty-eight hours later, focused regions of DNA damage were produced by using a scanning laser system. Cells were fixed for immunofluorescent staining by using antibodies to γH2AX (green) or suv39h1 (red). Nuclei were stained with DAPI (blue).
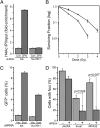
Fig. 2.
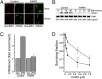
Suv39h1 regulates Tip60 activity, genomic stability, and homologous recombination (HR)-mediated repair. (A) 293T cells expressing nonspecific shRNA (control) or shRNA targeting suv39h1 were transfected with vector (Vec) or p84-ZFN (ZFN), followed by ChIP with antibody to H4Ac. RT-qPCR was carried out using primer pairs located to 1.5 kb to the right of the DSB. Results are ±SD (n = 3). (B) 293T cells expressing nonspecific shRNA (●) or shRNA targeting suv39h1 (○) were irradiated, and clonogenic cell survival assays were carried out. Results are ±SD (n = 3). (C) U2OS cells containing a GFP–HR reporter were stably transfected with nonspecific (NS) or shRNA targeting suv39h1, followed by transient transfection with vector (Vec) or the I-Sce1 endonuclease. GFP-positive cells were counted by FACS. Results are ±SD (n = 4 biological replicates). (D) U2OS cells expressing nonspecific (−) or shRNA targeting suv39h1 (+) were irradiated (2 Gy), allowed to recover for 30 min, and stained with antibodies to γH2AX, brca1, or RPA32. Cells with more than five foci were scored as positive. Results are ±SD (n = 3). P value was determined by using a t test.
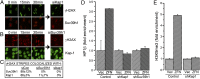
Fig. 3.
Recruitment of suv39h1 to DSBs requires kap-1 and HP1. (A) U2OS cells were transfected with control siRNA or siRNA to kap-1. DNA damage was created by using a laser, and cells were allowed to recover for 0, 15, or 30 min. Cells were costained with antibody to suv39h1 (red) or γH2AX (green). (B) U2OS cells were transfected with control siRNA or siRNA to suv39h1. DNA damage was created by using a laser, and cells were allowed to recover for 0, 15, or 30 min. Cells were costained with antibody to kap1 (green) or γH2AX (red). (C) Quantitation of results in A and B. The percentage of γH2AX laser stripes that colocalized with either suv39h1 or kap-1 stripes were noted. Results are ±SD (n = 25–60 cells). (D) 293T cells expressing nonspecific shRNA (control) or shRNA to kap-1 or suv39h1 were transfected with vector (Vec) or p84-ZFN (ZFN), followed by ChIP using HP1β antibody and primers 1.5 kb to the right of the DSB. Results are ±SD (n = 3). (E) 293T cells expressing nonspecific shRNA (control) or shRNA against kap-1 (shKap1) were transfected with p84-ZFN (ZFN) or vector (Vec) followed by ChIP using H3K9me3 antibody and primers located 1.5 kb to the right of the DSB. Results are ±SD (n = 3).
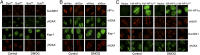
Fig. 4.
HP1’s chromodomain is required to recruit kap-1, HP1, and suv39h1 to DSBs. (A) U2OS cells expressing myc-suv39h1 (SuvWT) or catalytically inactive myc-suv39h1 (SuvMD) were exposed to laser damage and stained with antibody to myc and γH2AX or kap-1 and γH2AX. Some cells were preincubated in dimethyloxalylglycine (DMOG) (1 mM/1 h). (B) U2OS cells expressing nonspecific shRNA (shVec) or shRNA to suv39h1 (shSuv) were exposed to laser damage and costained with antibodies to HP1α and γH2AX or kap-1 and γH2AX. Some cells were preincubated in DMOG (1 mM/1 h). (C) U2OS cells expressing vector, HP1α (HA-HP1α), or HP1α with the chromodomain deleted (HA-HP1αCD) were exposed to laser damage and costained with antibodies for HA and γH2AX or suv39h1 and γH2AX. Some cells were preincubated in DMOG (1 mM/1 h).
Fig. 5.
Phosphorylation of kap-1 by ATM releases suv39h, kap-1, and HP1 from DSBs. (A and B) U2OS cells were preincubated with ATMi (100 μM) or solvent for 60 min. After laser microirradiation, cells were immediately fixed (0 min) or allowed to recover for 15 min and then costained with antibodies to suv39h1 and γH2AX (A) or kap-1 and γH2AX (B). (C) U2OS cells expressing wild-type kap-1 (mycKap1wt), kap-1 with an alanine mutation in the ATM phosphorylation site (mycKap1S824D), or kap-1 with a phospho-mimic in the same site (mycKap1S824D) were exposed to laser microirradiation and mycKap1 and γH2AX detected with myc or γH2AX antibody.
Fig. 6.
Chromatin PARylation recruits kap-1, HP1, and suv39h1 to DSBs. (A) U2OS cells were preincubated with PARPi (olaparib; 1 h/20 μM), followed by laser microirradiation. Cells were either fixed (0 min) or allowed to recover for 15 min, and then costained with antibody to suv39h1 and γH2AX. (B) U2OS cells were preincubated in PARPi (olaparib; 20 μM) for 1 h, and then exposed to bleomycin (5 μM) for the indicated times. Kap1, pkap1, and tubulin were monitored by Western blot analysis. (C) 293T cells were transfected with vector (Vec) or p84-ZFN (ZFN), followed by solvent (control) or PARPi (olaparib; 20μM). ChIP was carried out by using H3K9me3 antibody and primers located 1.5 kb to the right of the DSB. Results are ±SD (n = 3). (D) 293T cells expressing nonspecific shRNA (○) or shRNA targeting suv39h1 (●) were incubated with PARPi for 24 h, and clonogenic cell survival assays were carried out. Results are ±SE (n = 3).
Similar articles
- Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60.
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Sun Y, et al. Nat Cell Biol. 2009 Nov;11(11):1376-82. doi: 10.1038/ncb1982. Epub 2009 Sep 27. Nat Cell Biol. 2009. PMID: 19783983 Free PMC article. - PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination.
Clarke TL, Sanchez-Bailon MP, Chiang K, Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, Matias PM, Maslen SL, Skehel JM, Stewart GS, Davies CC. Clarke TL, et al. Mol Cell. 2017 Mar 2;65(5):900-916.e7. doi: 10.1016/j.molcel.2017.01.019. Epub 2017 Feb 23. Mol Cell. 2017. PMID: 28238654 Free PMC article. - Pericentric H3K9me3 Formation by HP1 Interaction-defective Histone Methyltransferase Suv39h1.
Muramatsu D, Kimura H, Kotoshiba K, Tachibana M, Shinkai Y. Muramatsu D, et al. Cell Struct Funct. 2016 Dec 3;41(2):145-152. doi: 10.1247/csf.16013. Epub 2016 Oct 12. Cell Struct Funct. 2016. PMID: 27733730 - Tip60: connecting chromatin to DNA damage signaling.
Sun Y, Jiang X, Price BD. Sun Y, et al. Cell Cycle. 2010 Mar 1;9(5):930-6. doi: 10.4161/cc.9.5.10931. Epub 2010 Mar 11. Cell Cycle. 2010. PMID: 20160506 Free PMC article. Review. - Mechanistic links between ATM and histone methylation codes during DNA repair.
Xu Y, Xu C, Price BD. Xu Y, et al. Prog Mol Biol Transl Sci. 2012;110:263-88. doi: 10.1016/B978-0-12-387665-2.00010-9. Prog Mol Biol Transl Sci. 2012. PMID: 22749149 Review.
Cited by
- Double-strand break repair and mis-repair in 3D.
Zagelbaum J, Gautier J. Zagelbaum J, et al. DNA Repair (Amst). 2023 Jan;121:103430. doi: 10.1016/j.dnarep.2022.103430. Epub 2022 Nov 17. DNA Repair (Amst). 2023. PMID: 36436496 Free PMC article. - Dynamic recruitment of UFM1-specific peptidase 2 to the DNA double-strand breaks regulated by WIP1.
Qin B, Yu J, Zhao F, Huang J, Zhou Q, Lou Z. Qin B, et al. Genome Instab Dis. 2022;3(4):217-226. doi: 10.1007/s42764-022-00076-z. Epub 2022 Aug 10. Genome Instab Dis. 2022. PMID: 36042814 Free PMC article. - Multi-Scale Imaging of the Dynamic Organization of Chromatin.
García Fernández F, Huet S, Miné-Hattab J. García Fernández F, et al. Int J Mol Sci. 2023 Nov 4;24(21):15975. doi: 10.3390/ijms242115975. Int J Mol Sci. 2023. PMID: 37958958 Free PMC article. Review. - Nascent Connections: R-Loops and Chromatin Patterning.
Chédin F. Chédin F. Trends Genet. 2016 Dec;32(12):828-838. doi: 10.1016/j.tig.2016.10.002. Epub 2016 Oct 25. Trends Genet. 2016. PMID: 27793359 Free PMC article. Review. - HDAC3 Deficiency Promotes Liver Cancer through a Defect in H3K9ac/H3K9me3 Transition.
Ji H, Zhou Y, Zhuang X, Zhu Y, Wu Z, Lu Y, Li S, Zeng Y, Lu QR, Huo Y, Shi Y, Bu H. Ji H, et al. Cancer Res. 2019 Jul 15;79(14):3676-3688. doi: 10.1158/0008-5472.CAN-18-3767. Epub 2019 May 16. Cancer Res. 2019. PMID: 31097476 Free PMC article.
References
- Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA93602/CA/NCI NIH HHS/United States
- R01 CA177884/CA/NCI NIH HHS/United States
- CA64585/CA/NCI NIH HHS/United States
- CA177884/CA/NCI NIH HHS/United States
- R01 CA064585/CA/NCI NIH HHS/United States
- R01 CA093602/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous