FUS is sequestered in nuclear aggregates in ALS patient fibroblasts - PubMed (original) (raw)
FUS is sequestered in nuclear aggregates in ALS patient fibroblasts
Jacob C Schwartz et al. Mol Biol Cell. 2014.
Abstract
Mutations in the RNA-binding protein FUS have been shown to cause the neurodegenerative disease amyotrophic lateral sclerosis (ALS). We investigate whether mutant FUS protein in ALS patient-derived fibroblasts affects normal FUS functions in the nucleus. We investigated fibroblasts from two ALS patients possessing different FUS mutations and a normal control. Fibroblasts from these patients have their nuclear FUS protein trapped in SDS-resistant aggregates. Genome-wide analysis reveals an inappropriate accumulation of Ser-2 phosphorylation on RNA polymerase II (RNA Pol II) near the transcription start sites of 625 genes for ALS patient cells and after small interfering RNA (siRNA) knockdown of FUS in normal fibroblasts. Furthermore, both the presence of mutant FUS protein and siRNA knockdown of wild-type FUS correlate with altered distribution of RNA Pol II within fibroblast nuclei. A loss of FUS function in orchestrating Ser-2 phosphorylation of the CTD of RNA Pol II is detectable in ALS patient-derived fibroblasts expressing mutant FUS protein, even when the FUS protein remains largely nuclear. A likely explanation for this loss of function is the aggregation of FUS protein in nuclei. Thus our results suggest a specific mechanism by which mutant FUS can have biological consequences other than by the formation of cytoplasmic aggregates.
© 2014 Schwartz et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
FIGURE 1:
ALS patient fibroblasts expressing mutant FUS show FUS predominantly in nuclei. (A) Three cell lines were tested. mFUS is homozygous for a point mutation, H517Q. ΔNLS is heterozygous for a frameshift mutation deleting the entire nuclear localization signal, M511Nfs. Wild type is a sex-matched normal control. (B) Immunofluorescence microscopy reveals that FUS is almost entirely nuclear, except for the ΔNLS cell line, which shows some cytoplasmic localization. Scale bar (lower left), 10 μm.
FIGURE 2:
Mutant FUS is trapped in aggregates within nuclei of ALS patient fibroblasts. (A) Western analysis of whole-cell FUS protein reveals that levels between the three cell lines are the same (WC, left). For ΔNLS samples, FUS protein from isolated nuclei after hypotonic lysis fails to resolve by SDS–PAGE (Nuclei, right). (B) Cytoplasmic FUS (Cyto., left) is also not detectable by SDS–PAGE. (C) Dot blot analysis of FUS protein isolated from cytoplasmic (C) or nuclear (N) fractions reveals that FUS is predominantly nuclear, in agreement with immunofluorescence data. (D) Treatment of nuclear protein with formic acid (Form., right) partially disrupts FUS aggregates, allowing protein to accumulate in the well during SDS–PAGE.
FIGURE 3:
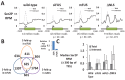
A subset of genes shows accumulation of Ser-2 phosphorylation (Ser2P) on RNA Pol II in ALS patient cells and after siRNA knockdown. (A) Average Ser2P signal for either all expressed genes (Total, dashed line) or the 625 genes at the intersect of B (Intersect, solid line) for wild-type cells, siFUS, mFUS, or ΔNLS cells. The p value is measured between normalized sum of Ser2P near the TSS for the intersect of treated compared with wild type and using the two-tailed Student's t test assuming unequal variances. (B) Using the threshold of twofold increase in Ser2P within 300 nucleotides of the transcription start site (TSS) with respect to wild-type cells, 625 genes show increased Ser2P in both ALS patient cells and after siRNA knockdown of FUS in wild-type cells (siFUS). See inset (right) for relative mRNA levels of FUS in siFUS-treated cells measured by real-time PCR. (C) Median Ser2P signal for either all expressed genes (Total) or the 625 genes at the intersect of B (Intersect). Error bars represent 25th and 75th percentiles. RPM, reads per million.
FIGURE 4:
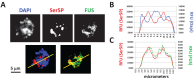
FUS and RNA Pol II immunofluorescence reveal their substantial colocalization and concentration in areas stained weakly by DAPI. (A) Nuclei stained with DAPI (stains DNA) and immunofluorescence for Ser5P (active RNA Pol II) and FUS. Both FUS and RNA Pol II show granular staining patterns within the nucleus of wild-type fibroblast cells. Yellow lines indicate line profiles quantified in B and C. (B) Quantification of relative fluorescence units (RFU) for Ser5P (red) and DAPI (blue) along the line in A, lower left. (C) Quantification of RFU for Ser5P (red) and FUS (green) along the line in A, lower right.
FIGURE 5:
RNA Pol II granules are more abundant and smaller in cells expressing mutant FUS or lacking FUS due to siRNA knockdown. (A–D) Four representative nuclei for wild-type, mutant FUS, or FUS knockdown. Scale bar (lower left), 5 μm. (E) The mean number of RNA Pol II-stained granules per nucleus is higher in mFUS, ΔNLS, and siFUS cells than in wild-type cells (n > 30 cells). Error bars, SD. **p < 0.0001, *_p_ < 0.05, Student's _t_ test. (F) The diameter of RNA Pol II granules at their widest cross section tends to be smaller in mFUS, ΔNLS, and siFUS cells than in wild-type cells (_n_ > 200 granules). As with E, p < 0.0001 for mFUS and ΔNLS vs. wild-type and p < 0.05 for siFUS vs. wild-type.
Similar articles
- Directly converted patient-specific induced neurons mirror the neuropathology of FUS with disrupted nuclear localization in amyotrophic lateral sclerosis.
Lim SM, Choi WJ, Oh KW, Xue Y, Choi JY, Kim SH, Nahm M, Kim YE, Lee J, Noh MY, Lee S, Hwang S, Ki CS, Fu XD, Kim SH. Lim SM, et al. Mol Neurodegener. 2016 Jan 22;11:8. doi: 10.1186/s13024-016-0075-6. Mol Neurodegener. 2016. PMID: 26795035 Free PMC article. - Mutations in the 3' untranslated region of FUS causing FUS overexpression are associated with amyotrophic lateral sclerosis.
Sabatelli M, Moncada A, Conte A, Lattante S, Marangi G, Luigetti M, Lucchini M, Mirabella M, Romano A, Del Grande A, Bisogni G, Doronzio PN, Rossini PM, Zollino M. Sabatelli M, et al. Hum Mol Genet. 2013 Dec 1;22(23):4748-55. doi: 10.1093/hmg/ddt328. Epub 2013 Jul 11. Hum Mol Genet. 2013. PMID: 23847048 - ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay.
Kamelgarn M, Chen J, Kuang L, Jin H, Kasarskis EJ, Zhu H. Kamelgarn M, et al. Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):E11904-E11913. doi: 10.1073/pnas.1810413115. Epub 2018 Nov 19. Proc Natl Acad Sci U S A. 2018. PMID: 30455313 Free PMC article. - TDP-43 and FUS en route from the nucleus to the cytoplasm.
Ederle H, Dormann D. Ederle H, et al. FEBS Lett. 2017 Jun;591(11):1489-1507. doi: 10.1002/1873-3468.12646. Epub 2017 Apr 23. FEBS Lett. 2017. PMID: 28380257 Review. - How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other?
Baloh RH. Baloh RH. Curr Opin Neurol. 2012 Dec;25(6):701-7. doi: 10.1097/WCO.0b013e32835a269b. Curr Opin Neurol. 2012. PMID: 23041957 Review.
Cited by
- Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins.
Lin Y, Protter DS, Rosen MK, Parker R. Lin Y, et al. Mol Cell. 2015 Oct 15;60(2):208-19. doi: 10.1016/j.molcel.2015.08.018. Epub 2015 Sep 24. Mol Cell. 2015. PMID: 26412307 Free PMC article. - Trends in Understanding the Pathological Roles of TDP-43 and FUS Proteins.
Buratti E. Buratti E. Adv Exp Med Biol. 2021;1281:243-267. doi: 10.1007/978-3-030-51140-1_15. Adv Exp Med Biol. 2021. PMID: 33433879 - The hnRNP RALY regulates PRMT1 expression and interacts with the ALS-linked protein FUS: implication for reciprocal cellular localization.
Gasperini L, Rossi A, Cornella N, Peroni D, Zuccotti P, Potrich V, Quattrone A, Macchi P. Gasperini L, et al. Mol Biol Cell. 2018 Dec 15;29(26):3067-3081. doi: 10.1091/mbc.E18-02-0108. Epub 2018 Oct 24. Mol Biol Cell. 2018. PMID: 30354839 Free PMC article. - m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress.
Yoneda R, Ueda N, Kurokawa R. Yoneda R, et al. Int J Mol Sci. 2021 Oct 12;22(20):11014. doi: 10.3390/ijms222011014. Int J Mol Sci. 2021. PMID: 34681673 Free PMC article. - Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions.
Farrawell NE, Lambert-Smith IA, Warraich ST, Blair IP, Saunders DN, Hatters DM, Yerbury JJ. Farrawell NE, et al. Sci Rep. 2015 Aug 21;5:13416. doi: 10.1038/srep13416. Sci Rep. 2015. PMID: 26293199 Free PMC article.
References
- Alliegro MC, Alliegro MA. A nuclear protein regulated during the transition from active to quiescent phenotype in cultured endothelial cells. Dev Biol. 1996;174:288–297. - PubMed
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM095311/GM/NIGMS NIH HHS/United States
- K99 NS082376/NS/NINDS NIH HHS/United States
- 1K99NS082376-01A1/NS/NINDS NIH HHS/United States
- 1F32GM095311-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous