Most genetic risk for autism resides with common variation - PubMed (original) (raw)
doi: 10.1038/ng.3039. Epub 2014 Jul 20.
Lambertus Klei 2, Stephan J Sanders 3, Corneliu A Bodea 1, Arthur P Goldberg 4, Ann B Lee 1, Milind Mahajan 5, Dina Manaa 5, Yudi Pawitan 6, Jennifer Reichert 7, Stephan Ripke 8, Sven Sandin 6, Pamela Sklar 9, Oscar Svantesson 6, Abraham Reichenberg 10, Christina M Hultman 6, Bernie Devlin 2, Kathryn Roeder 11, Joseph D Buxbaum 12
Affiliations
- PMID: 25038753
- PMCID: PMC4137411
- DOI: 10.1038/ng.3039
Most genetic risk for autism resides with common variation
Trent Gaugler et al. Nat Genet. 2014 Aug.
Abstract
A key component of genetic architecture is the allelic spectrum influencing trait variability. For autism spectrum disorder (herein termed autism), the nature of the allelic spectrum is uncertain. Individual risk-associated genes have been identified from rare variation, especially de novo mutations. From this evidence, one might conclude that rare variation dominates the allelic spectrum in autism, yet recent studies show that common variation, individually of small effect, has substantial impact en masse. At issue is how much of an impact relative to rare variation this common variation has. Using a unique epidemiological sample from Sweden, new methods that distinguish total narrow-sense heritability from that due to common variation and synthesis of results from other studies, we reach several conclusions about autism's genetic architecture: its narrow-sense heritability is ∼52.4%, with most due to common variation, and rare de novo mutations contribute substantially to individual liability, yet their contribution to variance in liability, 2.6%, is modest compared to that for heritable variation.
Figures
Figure 1
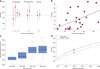
Results for PAGES (Population-based Autism Genetics and Environment Study), the Swedish study of the heritability of autism. (a) Heritability estimate (95% confidence interval) compared across study designs and analytical methods. Horizontal reference is the PAGES estimate of heritability from SNP genotypes. Twin studies: 1 California twins for strict autism (95% confidence interval: 8–84%), the largest twin study to date using diagnosis only; 2, Swedish twins 9–12 years old (95% confidence interval: 29–91%); 3, Swedish twins 9–12 years old characterized for a quantitative measure of autism (most extreme cutoff; 95% confidence interval: 44–74%). SNP-based estimates of heritability: 4, Swedish family study (95% confidence interval: 44–64%); 5, simplex cases versus population controls (95% confidence interval: 26–73%); 6, multiplex autism cases versus population controls (95% confidence interval: 38–93%). SNP-based estimates from the PAGES study, assuming prevalence K=0.3%; 7, heritability due to common variants using autism cases versus population controls (95% confidence interval: 31–69%); 8, total narrow-sense heritability due to both common and rare variation using smoothed estimates of relatedness (95% confidence interval: 35–71%). (b) Heritability per chromosome versus length in cM. (c) Prevalence by county for all 21 counties in Sweden tallied by birth year cohort. Each boxplot has a lower tail that extends from the minimum county-level prevalence to the 25th percentile; a central box that begins at the 25th percentile and ends at the 75th percentile, with a line demarcating the median prevalence; and an upper tail that extends from the 75th percentile to either (1) the maximum county-level prevalence (in the absence of any outliers) or (2) to a value of the 75th percentile + 1.5 times the vertical distance covered by the box – in this case, any outliers that exceed this end of the tail are noted by circular points on the plot. (d) PAGES heritability versus population prevalence of autism for two estimators of heritability: case-control contrast using SNP genotypes (green); total heritability from smoothed relationships amongst subjects, based on SNP genotypes (blue). Beyond the analysis of the PAGES study we applied meta-analysis of selected h2 estimates (Methods) to obtain h2 = 51.4% (SE = 5.2), which corresponds to a 95% confidence interval of (41.0, 61.8). Contrasting this with the comprehensive estimate of h2 obtained from the Swedish family study (h2 = 54%, SE = 5) produces an estimate of h2 due to rare variants: h2 = 2.6% (SE = 7.2, 95% confidence interval: 0–17%). Hence we conclude that common variants explain the bulk of the heritability for autism, at least 41% of the variability, and rare variants explain at most 17%, based on the upper and lower bounds of the respective 95% confidence intervals.
Figure 2
Results regarding the genetic architecture of autism spectrum disorder. The variance of autism liability is determined by genetic and environmental factors. The genetic factors include: ‘A’ additive effects, ‘D’ non-additive effects (dominant, recessive, epistatic), and ‘N’ de novo mutations. The environmental factors are split between ‘C’ common or shared environment and ‘E’ stochastic or unique environment. (a) Early autism twin studies estimate ‘A’ from the contrast of monozygotic (MZ) and dizygotic (DZ) correlations while assuming that ‘D’ and ‘N’ are zero. These are common assumptions for ‘ACE’ heritability models, but are unlikely to be appropriate for autism. (b) Applying the ACE model to the largest autism twin study to date yields a lower estimate of additive heritability. (c) Heritability results using a more extensive set of family relationships and based on much of the population of Sweden. (d) Results from the PAGES study (see Fig. 1). (e) Contribution of the various factors to the variance of autism liability according to family relationship. De novo variation should not be shared in dizygotic twins, and when it appears to be, it is almost surely inherited variation from a parent with gonadal mosaicism because the chance of the same mutation appearing de novo in the dizygotic twins is negligible. Most twin studies assume ‘C’ is the same for monozygotic and dizygotic twins, although that approximation has been debated. Of note, the excess covariance of monozygotic twins relative to dizygotic twins is 1/2 A+ 3/4 D + N as opposed to the 1/2 A assumed in the ACE model. (f) Synthesis of results for the genetic architecture of autism.
Comment in
- Common genetic variants linked with large percentage of autism risk: study finds spontaneous mutations are less-significant risk factors.
Levenson D. Levenson D. Am J Med Genet A. 2014 Nov;164A(11):vii-viii. doi: 10.1002/ajmg.a.36817. Am J Med Genet A. 2014. PMID: 25327469 No abstract available. - Autism genes: the continuum that connects us all.
Parihar R, Ganesh S. Parihar R, et al. J Genet. 2016 Sep;95(3):481-3. doi: 10.1007/s12041-016-0688-0. J Genet. 2016. PMID: 27659318 No abstract available.
Similar articles
- How rare and common risk variation jointly affect liability for autism spectrum disorder.
Klei L, McClain LL, Mahjani B, Panayidou K, De Rubeis S, Grahnat AS, Karlsson G, Lu Y, Melhem N, Xu X, Reichenberg A, Sandin S, Hultman CM, Buxbaum JD, Roeder K, Devlin B. Klei L, et al. Mol Autism. 2021 Oct 6;12(1):66. doi: 10.1186/s13229-021-00466-2. Mol Autism. 2021. PMID: 34615521 Free PMC article. - Analysis of common genetic variation and rare CNVs in the Australian Autism Biobank.
Yap CX, Alvares GA, Henders AK, Lin T, Wallace L, Farrelly A, McLaren T, Berry J, Vinkhuyzen AAE, Trzaskowski M, Zeng J, Yang Y, Cleary D, Grove R, Hafekost C, Harun A, Holdsworth H, Jellett R, Khan F, Lawson L, Leslie J, Levis Frenk M, Masi A, Mathew NE, Muniandy M, Nothard M, Visscher PM, Dawson PA, Dissanayake C, Eapen V, Heussler HS, Whitehouse AJO, Wray NR, Gratten J. Yap CX, et al. Mol Autism. 2021 Feb 10;12(1):12. doi: 10.1186/s13229-020-00407-5. Mol Autism. 2021. PMID: 33568206 Free PMC article. - Rhesus macaque social functioning is paternally, but not maternally, inherited by sons: potential implications for autism.
Garner JP, Talbot CF, Del Rosso LA, McCowan B, Kanthaswamy S, Haig D, Capitanio JP, Parker KJ. Garner JP, et al. Mol Autism. 2023 Jul 21;14(1):25. doi: 10.1186/s13229-023-00556-3. Mol Autism. 2023. PMID: 37480043 Free PMC article. - Rare variants and the oligogenic architecture of autism.
Wang T, Zhao PA, Eichler EE. Wang T, et al. Trends Genet. 2022 Sep;38(9):895-903. doi: 10.1016/j.tig.2022.03.009. Epub 2022 Apr 9. Trends Genet. 2022. PMID: 35410794 Free PMC article. Review. - The genetics of autism.
Muhle R, Trentacoste SV, Rapin I. Muhle R, et al. Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review.
Cited by
- Validity and reliability of chronic tic disorder and obsessive-compulsive disorder diagnoses in the Swedish National Patient Register.
Rück C, Larsson KJ, Lind K, Perez-Vigil A, Isomura K, Sariaslan A, Lichtenstein P, Mataix-Cols D. Rück C, et al. BMJ Open. 2015 Jun 22;5(6):e007520. doi: 10.1136/bmjopen-2014-007520. BMJ Open. 2015. PMID: 26100027 Free PMC article. - Quantitative genome-wide association study of six phenotypic subdomains identifies novel genome-wide significant variants in autism spectrum disorder.
Yousaf A, Waltes R, Haslinger D, Klauck SM, Duketis E, Sachse M, Voran A, Biscaldi M, Schulte-Rüther M, Cichon S, Nöthen M, Ackermann J, Koch I, Freitag CM, Chiocchetti AG. Yousaf A, et al. Transl Psychiatry. 2020 Jul 5;10(1):215. doi: 10.1038/s41398-020-00906-2. Transl Psychiatry. 2020. PMID: 32624584 Free PMC article. - Transcriptomic signatures of neuronal differentiation and their association with risk genes for autism spectrum and related neuropsychiatric disorders.
Chiocchetti AG, Haslinger D, Stein JL, de la Torre-Ubieta L, Cocchi E, Rothämel T, Lindlar S, Waltes R, Fulda S, Geschwind DH, Freitag CM. Chiocchetti AG, et al. Transl Psychiatry. 2016 Aug 2;6(8):e864. doi: 10.1038/tp.2016.119. Transl Psychiatry. 2016. PMID: 27483382 Free PMC article. - The road to precision psychiatry: translating genetics into disease mechanisms.
Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. Gandal MJ, et al. Nat Neurosci. 2016 Oct 26;19(11):1397-1407. doi: 10.1038/nn.4409. Nat Neurosci. 2016. PMID: 27786179 Free PMC article. Review. - The genetics and neurobiology of ESSENCE: The third Birgit Olsson lecture.
Bourgeron T. Bourgeron T. Nord J Psychiatry. 2016;70(1):1-9. doi: 10.3109/08039488.2015.1042519. Epub 2015 May 14. Nord J Psychiatry. 2016. PMID: 25971862 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- R01 MH095034/MH/NIMH NIH HHS/United States
- NS049261/NS/NINDS NIH HHS/United States
- R01 MH077139/MH/NIMH NIH HHS/United States
- CAPMC/ CIHR/Canada
- HD055784/HD/NICHD NIH HHS/United States
- MH057881/MH/NIMH NIH HHS/United States
- MH06359/MH/NIMH NIH HHS/United States
- HD055751/HD/NICHD NIH HHS/United States
- MH57881/MH/NIMH NIH HHS/United States
- MH077139/MH/NIMH NIH HHS/United States
- MH066673/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 MH097849/MH/NIMH NIH HHS/United States
- MH55284/MH/NIMH NIH HHS/United States
- MH080647/MH/NIMH NIH HHS/United States
- MH061009/MH/NIMH NIH HHS/United States
- MH095034/MH/NIMH NIH HHS/United States
- HD055782/HD/NICHD NIH HHS/United States
- MH081754/MH/NIMH NIH HHS/United States
- MH097849/MH/NIMH NIH HHS/United States
- R56 MH097849/MH/NIMH NIH HHS/United States
- MH66766/MH/NIMH NIH HHS/United States
- MH52708/MH/NIMH NIH HHS/United States
- R01 MH057881/MH/NIMH NIH HHS/United States
- NS042165/NS/NINDS NIH HHS/United States
- NS026630/NS/NINDS NIH HHS/United States
- HD35465/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials